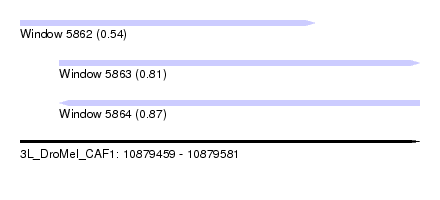
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,879,459 – 10,879,581 |
| Length | 122 |
| Max. P | 0.870290 |

| Location | 10,879,459 – 10,879,549 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -24.34 |
| Consensus MFE | -10.09 |
| Energy contribution | -10.62 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
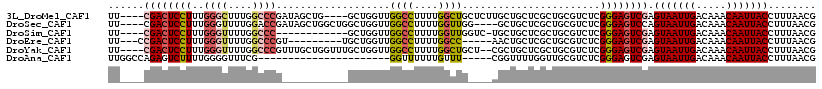
>3L_DroMel_CAF1 10879459 90 + 23771897 GGAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGCAAGAGCAGCCAAAAGGCCAA--CCAGC ((.....(((((((((....(((((((.....)))))))(((.....))))))))))))....((....)).(((....)))...--))... ( -27.90) >DroPse_CAF1 7711 84 + 1 GAAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUUGACUCCCGAGGAG---UCAGUCGC----ACAACCAG-AGCCCAAAACCAAA .......((.((((....(((((((((.....)))))))))((((((...))))---))...)))----))).....-.............. ( -21.10) >DroSec_CAF1 11779 86 + 1 GGAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUGGACUCCCGAGACGCAGCGAGCAGC----CCAACCAAAAGGCCAA--CCAGC ((.....(((((((((...((((((((.....))))))))((....))..)))))))))....((----(.........)))...--))... ( -27.00) >DroEre_CAF1 6915 85 + 1 GGAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGUU-----GGCCAAAAGGCCAA--CCAGC .......(((((((((....(((((((.....)))))))(((.....)))))))))))).((.(((-----((((....))))))--)..)) ( -34.10) >DroAna_CAF1 1824 78 + 1 GGAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAACCAAAACCG-----AAACAAAAAACC--------- ((........((((((....(((((((.....)))))))(((.....))))))))).......)).-----............--------- ( -14.86) >DroPer_CAF1 7231 84 + 1 GAAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUUGACUCCCGAGGAG---UCAGUCGC----ACAACCAG-AGCCCAAAACCAAA .......((.((((....(((((((((.....)))))))))((((((...))))---))...)))----))).....-.............. ( -21.10) >consensus GGAACAAUGCUGCGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCCAGCAGC_____CAACCAAAAGGCCAA__CCAGC ........(((((......((((((((.....))))))))...((....)).........)))))........................... (-10.09 = -10.62 + 0.53)



| Location | 10,879,471 – 10,879,581 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -16.85 |
| Energy contribution | -18.35 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10879471 110 + 23771897 CGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGCAAGAGCAGCCAAAAGGCCAACCAGC----CAGCUAUCGGGCCAAAGCCCAAAGGAGUCG----AA ........(((((((.....)))))))((((((((........(((..((.((....)).(((....)))......))----..)))...((((....))))...)))))))----). ( -38.30) >DroSec_CAF1 11791 110 + 1 CGUUAAAGGUAAUUGUUUGUCAAUUACUGGACUCCCGAGACGCAGCGAGCAGC----CCAACCAAAAGGCCAACCAGCCAGCCAGCUAUCGGUCCAAAACCCAAAGGAGUCG----AA .......((((((((.....)))))))).((((((.((...((.((.....))----..........(((......))).....))..))(((.....)))....)))))).----.. ( -27.10) >DroSim_CAF1 7822 101 + 1 CGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGCA-GACCAACCAAAAGGCCAACCAGC------------GGGCCAAAACCCAAAGGAGUCG----AA ........(((((((.....)))))))((((((((.........((.....)).-............((((.......------------.))))..........)))))))----). ( -29.20) >DroEre_CAF1 6927 101 + 1 CGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGUU-----GGCCAAAAGGCCAACCAGCA---------ACGGGCCAAAACCCAAAGGAGUCGG---AA .......((((((((.....))))))))(((((((.............((.(((-----((((....)))))))..)).---------..(((......)))...))))))).---.. ( -37.60) >DroYak_CAF1 7663 112 + 1 CGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGCG--AGCAGCCAAAAGGCCAACCAGCAAACCAGCAAACGGGCCAAAACCCAAAGGAGUCG----AA ........(((((((.....)))))))((((((((......((.((.....)).--.))........((((.....((......)).....))))..........)))))))----). ( -34.00) >DroAna_CAF1 1836 91 + 1 CGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAACCAAAACCG-----AAACAAAAAACC----------------------CGAAACCCCAAAAGACUCUGGCCAA .(((...((((((((.....))))).((((.....))))...........))).-----.)))........----------------------.......(((........))).... ( -11.30) >consensus CGUUAAAGGUAAUUGUUUGUCAAUUACUCGACUCCCGAGACGCAGCGAGCAGCA____CAACCAAAAGGCCAACCAGC___________CGGGCCAAAACCCAAAGGAGUCG____AA .......((((((((.....)))))))).((((((......((.....)).................((((....................))))..........))))))....... (-16.85 = -18.35 + 1.50)

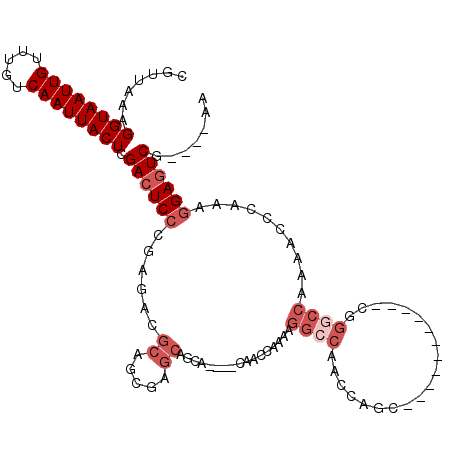

| Location | 10,879,471 – 10,879,581 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.84 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -21.19 |
| Energy contribution | -21.92 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10879471 110 - 23771897 UU----CGACUCCUUUGGGCUUUGGCCCGAUAGCUG----GCUGGUUGGCCUUUUGGCUGCUCUUGCUGCUCGCUGCGUCUCGGGAGUCGAGUAAUUGACAAACAAUUACCUUUAACG .(----((((((((((((((....))))))((((.(----((.....((((....))))((....)).))).))))......)))))))))(((((((.....)))))))........ ( -43.70) >DroSec_CAF1 11791 110 - 1 UU----CGACUCCUUUGGGUUUUGGACCGAUAGCUGGCUGGCUGGUUGGCCUUUUGGUUGG----GCUGCUCGCUGCGUCUCGGGAGUCCAGUAAUUGACAAACAAUUACCUUUAACG ..----.(((((((...((((...))))(((.((.(((.(((.(.(..(((....)))..)----.).))).))))))))..)))))))..(((((((.....)))))))........ ( -37.70) >DroSim_CAF1 7822 101 - 1 UU----CGACUCCUUUGGGUUUUGGCCC------------GCUGGUUGGCCUUUUGGUUGGUC-UGCUGCUCGCUGCGUCUCGGGAGUCGAGUAAUUGACAAACAAUUACCUUUAACG .(----((((((((..((((....))))------------((.((..((((....))))..))-.)).((.....)).....)))))))))(((((((.....)))))))........ ( -36.90) >DroEre_CAF1 6927 101 - 1 UU---CCGACUCCUUUGGGUUUUGGCCCGU---------UGCUGGUUGGCCUUUUGGCC-----AACUGCUCGCUGCGUCUCGGGAGUCGAGUAAUUGACAAACAAUUACCUUUAACG ..---.((((((((..((((....))))((---------.(((((((((((....))))-----)))))...)).)).....)))))))).(((((((.....)))))))........ ( -40.90) >DroYak_CAF1 7663 112 - 1 UU----CGACUCCUUUGGGUUUUGGCCCGUUUGCUGGUUUGCUGGUUGGCCUUUUGGCUGCU--CGCUGCUCGCUGCGUCUCGGGAGUCGAGUAAUUGACAAACAAUUACCUUUAACG .(----((((((((..((((....))))((..((.(((..((.(((.((((....)))))))--.)).))).)).)).....)))))))))(((((((.....)))))))........ ( -41.30) >DroAna_CAF1 1836 91 - 1 UUGGCCAGAGUCUUUUGGGGUUUCG----------------------GGUUUUUUGUUU-----CGGUUUUGGUUGCGUCUCGGGAGUCGAGUAAUUGACAAACAAUUACCUUUAACG ....(((((....))))).(((..(----------------------(((...((((((-----...((((((.......))))))(((((....)))))))))))..))))..))). ( -19.80) >consensus UU____CGACUCCUUUGGGUUUUGGCCCG___________GCUGGUUGGCCUUUUGGCUG____CGCUGCUCGCUGCGUCUCGGGAGUCGAGUAAUUGACAAACAAUUACCUUUAACG ......((((((((..((((....))))...................((((....)))).......................)))))))).(((((((.....)))))))........ (-21.19 = -21.92 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:58 2006