
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,874,076 – 10,874,172 |
| Length | 96 |
| Max. P | 0.807370 |

| Location | 10,874,076 – 10,874,172 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.71 |
| Mean single sequence MFE | -21.83 |
| Consensus MFE | -14.79 |
| Energy contribution | -15.15 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.807370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
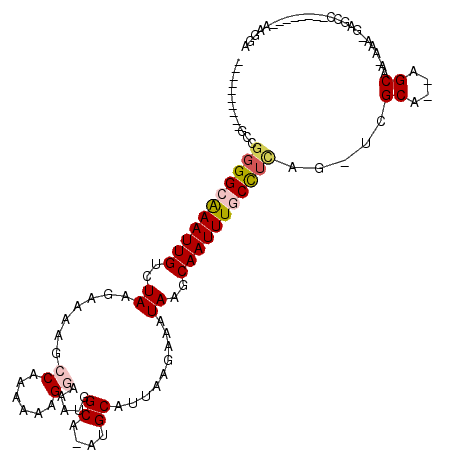
>3L_DroMel_CAF1 10874076 96 - 23771897 ----------GCCGGGGCAAAUUGUCUAAGAAAAGCCAAAAAAGGAAAUGGCA-AUGCAUUAAGAAAUAAGCAAUUUGCCUCAG-UCGCA--AGCAA-AAA-GAGCC--------AAGCA ----------((.((((((((((((.........((((..........)))).-........(....)..)))))))))))).(-.(...--.))..-...-.....--------..)). ( -23.20) >DroPse_CAF1 2995 117 - 1 UUG--GGGGGGACGGGGCGAAUUGUCUAAGAAAAGACAAAAAAGGAAAUGGCAAAUGCAUUAAGAAAUAAGCAAUUUUCUUUUGCUUGCAACAGCAA-AAACGUGGCACGAGAUCAAGGA (((--...(..(((((((...((((((......))))))((((((((((.((..................)).))))))))))))))((....))..-...)))..).)))......... ( -20.97) >DroSim_CAF1 2036 96 - 1 ----------GCCGGGGCAAAUUGUCUAAGAAAAGCCAAAAAAGGAAAUGGCA-AUGCAUUAAGAAAUAAGCAAUUUGCUUCAG-UCGCA--AGCAA-AAA-GAGCC--------AAGCA ----------((.((((((((((((.........((((..........)))).-........(....)..)))))))))))).(-.(...--.))..-...-.....--------..)). ( -20.40) >DroEre_CAF1 1466 95 - 1 ----------GCCC-GGCAAAUUGUCUAAGAAAAGCCAAAAAAGGAAAUGGCA-AUGCAUUAAGAAAUAAGCAAUUUGCCUCAG-ACGCA--AGCAAAAAG-GAGCC---------CGUA ----------((((-((((((((((.........((((..........)))).-........(....)..))))))))))...(-.(...--.)).....)-).)).---------.... ( -21.90) >DroYak_CAF1 1945 95 - 1 ----------GCCGGGGCAAAUUGUCUAAGAAAAGCCAAAAAAGGAAAUGGCA-AUGCAUUAAGAAAUAAGCAAUUUGCCUCAG-UCGCA--AGCAA-AAG-AAGCC---------AGGA ----------.((((((((((((((.........((((..........)))).-........(....)..)))))))))))).(-.(...--.))..-...-.....---------.)). ( -23.60) >DroPer_CAF1 2477 119 - 1 UUAGGGGGGGGACGGGGCGAAUUGUCUAAGAAAAGACAAAAAAGGAAAUGGCAAAUGCAUUAAGAAAUAAGCAAUUUUCUUUUGCUUGCAACAGCAA-AAACGUGGCACGAGAUCAAGGA .......(....)((..((..((((((......)))))).........((.((...(((..((((((........)))))).)))((((....))))-.....)).))))...))..... ( -20.90) >consensus __________GCCGGGGCAAAUUGUCUAAGAAAAGCCAAAAAAGGAAAUGGCA_AUGCAUUAAGAAAUAAGCAAUUUGCCUCAG_UCGCA__AGCAA_AAA_GAGCC________AAGGA .............(((((((((((..((.......((......)).....((....)).........))..))))))))))).....((....))......................... (-14.79 = -15.15 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:55 2006