
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
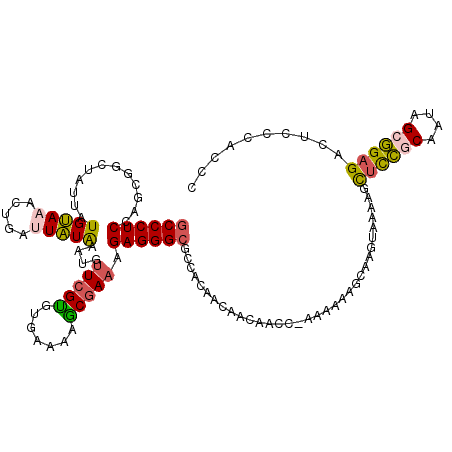
| Location | 10,873,288 – 10,873,431 |
| Length | 143 |
| Max. P | 0.740618 |

| Location | 10,873,288 – 10,873,391 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.46 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10873288 103 - 23771897 GCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUGAAUGUUCGUGUGAAAAACAAAAGAGGGCGCCACUACAACA-----------------AAGGUUUGGCGAUAGCCGAGACUCCCACCC (((((((((..((......))...)))..(((((....)))))............))))))............-----------------..((((((((....))).)))))....... ( -24.30) >DroSec_CAF1 1246 116 - 1 GCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUAA--GGUCGUGUGAAAAGCGAAAGAGGGCGCCACAGCAACAACC-AAAAAAGCAAGUAAAAGCUCCGCUUUAGCGGAGAC-CCCACCC ((((((..((((((.(((((.........)))))--))))))((.....))....))))))((....)).......-.................((((((....))))))..-....... ( -31.10) >DroSim_CAF1 1231 119 - 1 GCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUAAAUGUUCGCGUGAAAAGCGAAAGAGGGCGCCGCAACAACAACC-AAAAAAGCAAGUAAAAGCUCCGCAUUAGCGGAGACUCCCACCC (((((((((..((......))...)))..........(((((.......))))).))))))...............-...............((((((((....)))))).))....... ( -32.40) >DroEre_CAF1 615 120 - 1 ACCCUCUAGUGUCUAUCCUGCAAACUCGGUGUAAGUGUUCGUCACUUAAACGAAAGAGGGCGCCACAGCACAAACCUAGGAAAUCAGCUACAAGCUCCACAAUAGCGGAGACUCUCACCC ......((((.....((((((...(((.((.((((((.....)))))).))....))).))((....))........)))).....))))..((((((.(....).)))).))....... ( -25.20) >DroYak_CAF1 1093 120 - 1 GCCCUCUAGUGCCUGUCAUGCAAACUUGGUGUAAAUGUUCGCGACGAAAGCGAAAGAGGGCGCCACAACAACACCCUAAAGAAGCAAGUACAAGCACCGCAAUAGGGGAGACUUCCACCC ..((((((((((.((((.(((.....((((((.....(((((.......))))).....))))))..........(....)..))).).))).)))).....))))))............ ( -31.60) >consensus GCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUAAAUGUUCGUGUGAAAAGCGAAAGAGGGCGCCACAACAACAACC_AAAAAAGCAAGUAAAAGCUCCGCAAUAGCGGAGACUCCCACCC ((((((............(((((.....)))))....(((((.......))))).)))))).................................((((((....)))))).......... (-21.62 = -21.46 + -0.16)

| Location | 10,873,318 – 10,873,431 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.60 |
| Mean single sequence MFE | -33.28 |
| Consensus MFE | -20.14 |
| Energy contribution | -19.26 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10873318 113 + 23771897 -------UGUUGUAGUGGCGCCCUCUUUUGUUUUUCACACGAACAUUCAUAAUCAGUUUACAUAAUAGCCGCUGGAGGGCGCUCAUGUAUAUUUCCAGAUCCAGUACAAUGCGACUAAAG -------.(((((((((((((((((...(((((.......)))))........((((.............)))))))))))).)))((((.............))))..))))))..... ( -28.74) >DroSec_CAF1 1285 117 + 1 UUU-GGUUGUUGCUGUGGCGCCCUCUUUCGCUUUUCACACGACC--UUAUAAUCAGUUUACAUAAUAGCCGCUGGAGGGCGCUCAUGUAUAUUUCAAGAUCCAAUACUAUGGGAAUAAAA (((-((....(((.((((((((((((..((((........(((.--.........)))........)).))..))))))))).))))))....))))).((((......))))....... ( -29.29) >DroSim_CAF1 1271 112 + 1 UUU-GGUUGUUGUUGCGGCGCCCUCUUUCGCUUUUCACGCGAACAUUUAUAAUCAGUUUACAUAAUAGCCGCUGGAGGGCGCUCAUGUAUAUUUCAAGAUCCAGUACUA-------AAAA .((-((.(.(((.((((((((((((.(((((.......)))))..........((((.............)))))))))))))...))).....))).).)))).....-------.... ( -30.42) >DroEre_CAF1 655 120 + 1 CCUAGGUUUGUGCUGUGGCGCCCUCUUUCGUUUAAGUGACGAACACUUACACCGAGUUUGCAGGAUAGACACUAGAGGGUGCUCAGGUAUAUUUAAAGUUCCAUUGCCGCCAGACUGCGA ....((((((.((((.((((((((((.(((..((((((.....))))))...)))(((((.....)))))...))))))))))))((((...............)))))))))))).... ( -37.36) >DroYak_CAF1 1133 120 + 1 UUUAGGGUGUUGUUGUGGCGCCCUCUUUCGCUUUCGUCGCGAACAUUUACACCAAGUUUGCAUGACAGGCACUAGAGGGCGCACAAGUGUAUUUAAACGUCUUGUAUUGUUAGGCUGAAA (((((............(((((((((...((((((((.((((((...........)))))))))).))))...)))))))))((((((((......))).))))).........))))). ( -40.60) >consensus UUU_GGUUGUUGUUGUGGCGCCCUCUUUCGCUUUUCACACGAACAUUUAUAAUCAGUUUACAUAAUAGCCGCUGGAGGGCGCUCAUGUAUAUUUCAAGAUCCAGUACUAUGAGACUAAAA .................((((((((((((((.......)))))............(((.....))).......)))))))))...................................... (-20.14 = -19.26 + -0.88)


| Location | 10,873,318 – 10,873,431 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.60 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.34 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10873318 113 - 23771897 CUUUAGUCGCAUUGUACUGGAUCUGGAAAUAUACAUGAGCGCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUGAAUGUUCGUGUGAAAAACAAAAGAGGGCGCCACUACAACA------- ...........(((((.((..((((........)).))(((((((((((..((......))...)))..(((((....)))))............)))))))))).)))))..------- ( -26.10) >DroSec_CAF1 1285 117 - 1 UUUUAUUCCCAUAGUAUUGGAUCUUGAAAUAUACAUGAGCGCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUAA--GGUCGUGUGAAAAGCGAAAGAGGGCGCCACAGCAACAACC-AAA (((((...(((......)))....)))))......((.((((((((..((((((.(((((.........)))))--))))))((.....))....))))))))))...........-... ( -27.90) >DroSim_CAF1 1271 112 - 1 UUUU-------UAGUACUGGAUCUUGAAAUAUACAUGAGCGCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUAAAUGUUCGCGUGAAAAGCGAAAGAGGGCGCCGCAACAACAACC-AAA ....-------......(((.((.((.......)).))(((((((((((..((......))...)))..........(((((.......))))).))))))))...........))-).. ( -28.40) >DroEre_CAF1 655 120 - 1 UCGCAGUCUGGCGGCAAUGGAACUUUAAAUAUACCUGAGCACCCUCUAGUGUCUAUCCUGCAAACUCGGUGUAAGUGUUCGUCACUUAAACGAAAGAGGGCGCCACAGCACAAACCUAGG ..((....(((((((...((.............))...)).((((((..(((.......)))...(((...((((((.....))))))..))).)))))))))))..))........... ( -29.32) >DroYak_CAF1 1133 120 - 1 UUUCAGCCUAACAAUACAAGACGUUUAAAUACACUUGUGCGCCCUCUAGUGCCUGUCAUGCAAACUUGGUGUAAAUGUUCGCGACGAAAGCGAAAGAGGGCGCCACAACAACACCCUAAA ...............(((((..((......)).)))))(((((((((.(..((.((.......))..))..).....(((((.......))))))))))))))................. ( -30.70) >consensus UUUUAGUCUCAUAGUACUGGAUCUUGAAAUAUACAUGAGCGCCCUCCAGCGGCUAUUAUGUAAACUGAUUAUAAAUGUUCGUGUGAAAAGCGAAAGAGGGCGCCACAACAACAACC_AAA ...................................((.((((((((............(((((.....)))))....(((((.......))))).))))))))))............... (-20.62 = -20.34 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:54 2006