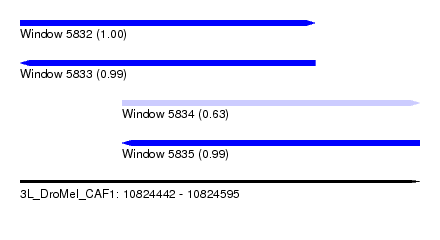
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,824,442 – 10,824,595 |
| Length | 153 |
| Max. P | 0.999817 |

| Location | 10,824,442 – 10,824,555 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 91.56 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -29.98 |
| Energy contribution | -29.82 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.15 |
| SVM RNA-class probability | 0.999817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10824442 113 + 23771897 CGGGUAACUGCAUGAAUAAUUAUUAAGUUAUUCGAGGAGGGUCGUAAAAUGAGGGGUAUGCAGAUGUACUGCACUCACUCGCACUCACCCCAAAAAUUUAUGGGAUUUCUCCC (((((((((..((((....))))..))))))))).((((((.........(((((((..((((.....)))))))).)))........((((........))))..)))))). ( -31.40) >DroSec_CAF1 33493 112 + 1 CGGGUAACUGCAUGAAUAAUUAUUAAGUUAUUCGAGGAGGGUCAUAAAAUGAGGGGUAUGCAGAAGUACUGCACUCACUCGCACUCACCCCAA-AAUUUAUGGGUUUUCUCCC .((((((((..((((....))))..))))))))(((((((.(((((((.(..(((((.(((((.....)))))(......).....)))))..-).))))))).))))))).. ( -32.70) >DroSim_CAF1 35101 112 + 1 CGGGUAACUGCAUGAAUAAUUAUUAAGUUAUUCGAGGAGGGUCAUAAAAUGAGGGGUAUGCAGAAGUACUGCACUCACUCGCACUCACCCCAA-AAUUUAUGGGUUUUCUCCC .((((((((..((((....))))..))))))))(((((((.(((((((.(..(((((.(((((.....)))))(......).....)))))..-).))))))).))))))).. ( -32.70) >DroEre_CAF1 34881 102 + 1 CGGGUAACUGCAUGAAUAAUUAUUAAGUUAUUCGAGGAGGGUCAUAAAACGAGGGGUAUGCAGAAGUGCUGCACU----------CACCCCAA-AAUUUAUGGGAUUUCUCCC .((((((((..((((....))))..))))))))((((((..(((((((....(((((.(((((.....)))))..----------.)))))..-..)))))))..)))))).. ( -33.70) >DroYak_CAF1 34189 102 + 1 CGGGUAACUGCAUGAAUAAUUAUUAAGUUAUUCGAGGAGGGUCAUAAAAUAGGGGGUAUGCAGAAGUACUGCACU----------CACCCCAA-AAUUUAUGGGAUUUCUCCC .((((((((..((((....))))..))))))))((((((..(((((((....(((((.(((((.....)))))..----------.)))))..-..)))))))..)))))).. ( -34.60) >consensus CGGGUAACUGCAUGAAUAAUUAUUAAGUUAUUCGAGGAGGGUCAUAAAAUGAGGGGUAUGCAGAAGUACUGCACUCACUCGCACUCACCCCAA_AAUUUAUGGGAUUUCUCCC (((((((((..((((....))))..))))))))).(((((((((((((....(((((.(((((.....))))).............))))).....)))))))...)))))). (-29.98 = -29.82 + -0.16)

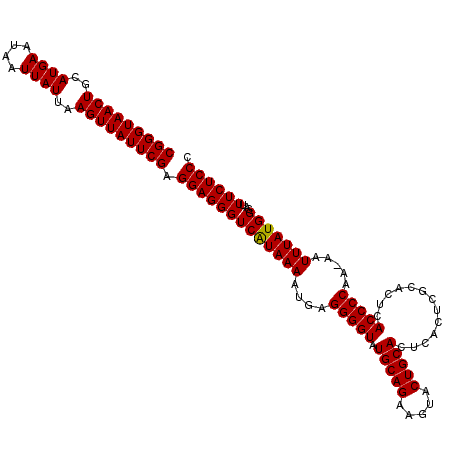

| Location | 10,824,442 – 10,824,555 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 91.56 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -25.76 |
| Energy contribution | -27.76 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10824442 113 - 23771897 GGGAGAAAUCCCAUAAAUUUUUGGGGUGAGUGCGAGUGAGUGCAGUACAUCUGCAUACCCCUCAUUUUACGACCCUCCUCGAAUAACUUAAUAAUUAUUCAUGCAGUUACCCG ((((....))))...........((((((.(((((((((((((((.....))))).....))))))).............((((((........))))))..))).)))))). ( -38.00) >DroSec_CAF1 33493 112 - 1 GGGAGAAAACCCAUAAAUU-UUGGGGUGAGUGCGAGUGAGUGCAGUACUUCUGCAUACCCCUCAUUUUAUGACCCUCCUCGAAUAACUUAAUAAUUAUUCAUGCAGUUACCCG (((......))).......-...((((((.((((((.((((((((.....)))))......((((...))))..))))))((((((........))))))..))).)))))). ( -35.10) >DroSim_CAF1 35101 112 - 1 GGGAGAAAACCCAUAAAUU-UUGGGGUGAGUGCGAGUGAGUGCAGUACUUCUGCAUACCCCUCAUUUUAUGACCCUCCUCGAAUAACUUAAUAAUUAUUCAUGCAGUUACCCG (((......))).......-...((((((.((((((.((((((((.....)))))......((((...))))..))))))((((((........))))))..))).)))))). ( -35.10) >DroEre_CAF1 34881 102 - 1 GGGAGAAAUCCCAUAAAUU-UUGGGGUG----------AGUGCAGCACUUCUGCAUACCCCUCGUUUUAUGACCCUCCUCGAAUAACUUAAUAAUUAUUCAUGCAGUUACCCG ((((....)))).......-...(((((----------(.(((((((....)))..........................((((((........)))))).)))).)))))). ( -30.10) >DroYak_CAF1 34189 102 - 1 GGGAGAAAUCCCAUAAAUU-UUGGGGUG----------AGUGCAGUACUUCUGCAUACCCCCUAUUUUAUGACCCUCCUCGAAUAACUUAAUAAUUAUUCAUGCAGUUACCCG (((........((((((..-..((((.(----------.((((((.....)))))).)))))...)))))).........((((((........)))))).........))). ( -29.40) >consensus GGGAGAAAUCCCAUAAAUU_UUGGGGUGAGUGCGAGUGAGUGCAGUACUUCUGCAUACCCCUCAUUUUAUGACCCUCCUCGAAUAACUUAAUAAUUAUUCAUGCAGUUACCCG ((((....))))...........((((((.(((......((((((.....))))))........................((((((........))))))..))).)))))). (-25.76 = -27.76 + 2.00)


| Location | 10,824,481 – 10,824,595 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -22.28 |
| Energy contribution | -22.36 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10824481 114 + 23771897 GGUCGUAAAAUGAGGGGUAUGCAGAUGUACUGCACUCACUCGCACUCACCCCAAAAAUUUAUGGGAUUUCUCCCUGAUAUCCGACUUAUUUUUAGACGUGCUCAAUCAUUUCUU .......((((((((((..(((((.....)))))..).)))((((((.....((((((...(.((((.((.....)).)))).)...)))))).)).))))....))))))... ( -24.90) >DroSec_CAF1 33532 113 + 1 GGUCAUAAAAUGAGGGGUAUGCAGAAGUACUGCACUCACUCGCACUCACCCCAA-AAUUUAUGGGUUUUCUCCCUGAUAUCCGACUUAUUUUUAGACGUGCUCAAUCAUUUCUU .(((..((((((((((((((.(((......(((........))).....((((.-......))))........)))))))))..))))))))..)))................. ( -23.20) >DroSim_CAF1 35140 113 + 1 GGUCAUAAAAUGAGGGGUAUGCAGAAGUACUGCACUCACUCGCACUCACCCCAA-AAUUUAUGGGUUUUCUCCCUGAUAUCCGACUUAUUUUUAGACGUGCUCAAUCAUUUCUU .(((..((((((((((((((.(((......(((........))).....((((.-......))))........)))))))))..))))))))..)))................. ( -23.20) >DroEre_CAF1 34920 103 + 1 GGUCAUAAAACGAGGGGUAUGCAGAAGUGCUGCACU----------CACCCCAA-AAUUUAUGGGAUUUCUCCCUGAUAUCCGACUUAUUUUUAGACGUGCUCAAUCAUUUCUU ((((.........(((((.(((((.....)))))..----------.)))))..-........((((.((.....)).)))))))).......(((.(((......))).))). ( -26.00) >DroYak_CAF1 34228 103 + 1 GGUCAUAAAAUAGGGGGUAUGCAGAAGUACUGCACU----------CACCCCAA-AAUUUAUGGGAUUUCUCCCUGAUAUCCGACUUAUUUUUAAACGUGCUCAAUCAUUUCUU (((((((((....(((((.(((((.....)))))..----------.)))))..-..))))).((((.((.....)).))))))))............................ ( -25.50) >consensus GGUCAUAAAAUGAGGGGUAUGCAGAAGUACUGCACUCACUCGCACUCACCCCAA_AAUUUAUGGGAUUUCUCCCUGAUAUCCGACUUAUUUUUAGACGUGCUCAAUCAUUUCUU .(((..((((((((((((((((((.....))))................((((........))))...........))))))..))))))))..)))................. (-22.28 = -22.36 + 0.08)

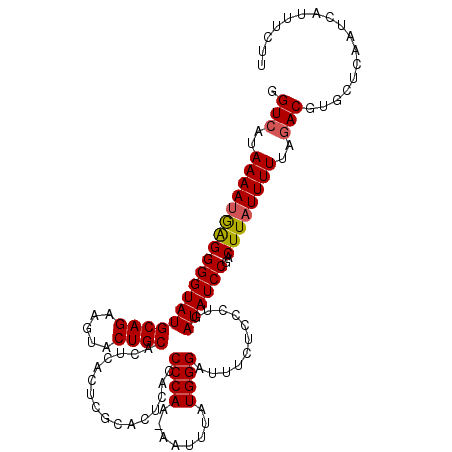
| Location | 10,824,481 – 10,824,595 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 91.28 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.54 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.992422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10824481 114 - 23771897 AAGAAAUGAUUGAGCACGUCUAAAAAUAAGUCGGAUAUCAGGGAGAAAUCCCAUAAAUUUUUGGGGUGAGUGCGAGUGAGUGCAGUACAUCUGCAUACCCCUCAUUUUACGACC ..(((((((.(((...((.((.......)).))....)))((((....))))..........((((((...((......))((((.....)))).)))))))))))))...... ( -31.60) >DroSec_CAF1 33532 113 - 1 AAGAAAUGAUUGAGCACGUCUAAAAAUAAGUCGGAUAUCAGGGAGAAAACCCAUAAAUU-UUGGGGUGAGUGCGAGUGAGUGCAGUACUUCUGCAUACCCCUCAUUUUAUGACC ..(((((((.(((...((.((.......)).))....)))(((......))).......-..((((((...((......))((((.....)))).)))))))))))))...... ( -27.40) >DroSim_CAF1 35140 113 - 1 AAGAAAUGAUUGAGCACGUCUAAAAAUAAGUCGGAUAUCAGGGAGAAAACCCAUAAAUU-UUGGGGUGAGUGCGAGUGAGUGCAGUACUUCUGCAUACCCCUCAUUUUAUGACC ..(((((((.(((...((.((.......)).))....)))(((......))).......-..((((((...((......))((((.....)))).)))))))))))))...... ( -27.40) >DroEre_CAF1 34920 103 - 1 AAGAAAUGAUUGAGCACGUCUAAAAAUAAGUCGGAUAUCAGGGAGAAAUCCCAUAAAUU-UUGGGGUG----------AGUGCAGCACUUCUGCAUACCCCUCGUUUUAUGACC ..(((((((.(((...((.((.......)).))....)))((((....)))).......-..(((((.----------.((((((.....))))))))))))))))))...... ( -31.50) >DroYak_CAF1 34228 103 - 1 AAGAAAUGAUUGAGCACGUUUAAAAAUAAGUCGGAUAUCAGGGAGAAAUCCCAUAAAUU-UUGGGGUG----------AGUGCAGUACUUCUGCAUACCCCCUAUUUUAUGACC ...(((((........)))))........(((........((((....))))(((((..-..((((.(----------.((((((.....)))))).)))))...)))))))). ( -29.80) >consensus AAGAAAUGAUUGAGCACGUCUAAAAAUAAGUCGGAUAUCAGGGAGAAAUCCCAUAAAUU_UUGGGGUGAGUGCGAGUGAGUGCAGUACUUCUGCAUACCCCUCAUUUUAUGACC ..(((((((.(((...((.((.......)).))....)))((((....))))..........(((((............((((((.....))))))))))))))))))...... (-26.42 = -26.54 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:30 2006