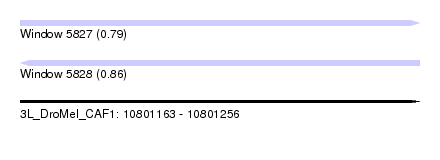
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,801,163 – 10,801,256 |
| Length | 93 |
| Max. P | 0.863063 |

| Location | 10,801,163 – 10,801,256 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.25 |
| Mean single sequence MFE | -19.55 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.77 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
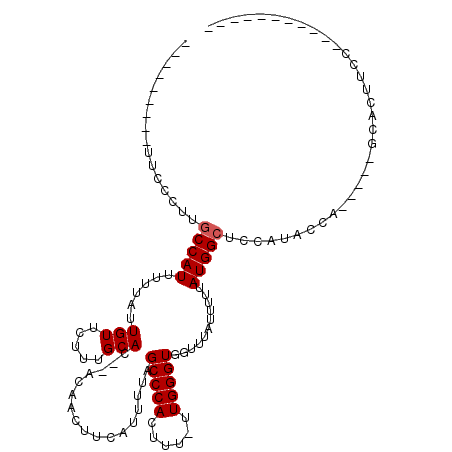
>3L_DroMel_CAF1 10801163 93 + 23771897 ---------UUCCUUUGCCAUUUUUAUUGUUCUUGGCACACACAACUUCAUUUUAGCCCACUU--UUGGGUGGUUUAUUUUUAUGGCUCCAUCCCC-----AAAGUUCC----------- ---------......(((((.............))))).....................((((--(.((((((.((((....))))..))).))).-----)))))...----------- ( -16.42) >DroVir_CAF1 28246 106 + 1 CGUGCGUCCUCUGCCUGCCAUUUUUAUUGUUCUUUGCAC--AGAACUUCAUUUUAGCCCACUUU-UUGGGUGGUUUAUUUUUAUGGCUCCAUAUCAUGCCCGCACCGCC----------- .(((((..........(((.........(((((......--))))).........(((((....-.))))))))..........(((..........))))))))....----------- ( -23.00) >DroPse_CAF1 91589 105 + 1 ---------UGCCUUUACCAUUUUUAUUGUUCUUCGCACACACAACUUCAUUUUAGCCCACUUU-UUGGGUGGUUUAUUUUUAUGGCUCCAUCCCU-----GGAGGAGCCCCCGCCCAGC ---------(((......((.......))......)))..........................-((((((((...........((((((.((...-----.)))))))).)))))))). ( -23.60) >DroGri_CAF1 28559 94 + 1 ------------GCCUGCCAUUUUUAUUGUUCUUUGCAC--ACAACUUCAUUUUAGCCCACUUUUUUGGGUGGCUUAUUUGUAUGGCUCCAUAUCAUGACG-CCCUUAC----------- ------------....(((........(((.....))).--..............(((((......))))))))......(((.(((((........)).)-))..)))----------- ( -15.50) >DroMoj_CAF1 23468 96 + 1 ---------CUGGCUUGCCAUUUUUAUUGUUCUUUGCAC--ACAACUUCAUUGUAGCCCACUUU-UUGGGUGGUUUAUUUUUAUGGCUCCAUAUCAGGGCC-CUCUGCC----------- ---------..(((..(((....................--((((.....)))).(((((....-.))))))))..........(((.((......)))))-....)))----------- ( -18.90) >DroAna_CAF1 5076 104 + 1 ---------UUCCUUUGCCAUUUUUAUUGUUCUUGGCACACACAACUUCAUUUUAGCCCACUUU-UUGGGUGGUUUAUUUUUAUGGCUCCAUCCCU-----GAAUUUGUCCC-GGCGAGC ---------....((((((....................................(((((....-.)))))((..((..((((.((......)).)-----)))..))..))-)))))). ( -19.90) >consensus _________UUCCCUUGCCAUUUUUAUUGUUCUUUGCAC__ACAACUUCAUUUUAGCCCACUUU_UUGGGUGGUUUAUUUUUAUGGCUCCAUACCA_____GCACUUCC___________ ................(((((......(((.....))).................(((((......)))))...........)))))................................. (-11.60 = -11.77 + 0.17)


| Location | 10,801,163 – 10,801,256 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.25 |
| Mean single sequence MFE | -22.26 |
| Consensus MFE | -12.52 |
| Energy contribution | -12.55 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
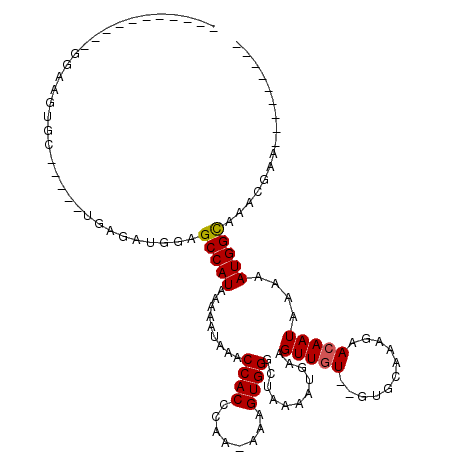
>3L_DroMel_CAF1 10801163 93 - 23771897 -----------GGAACUUU-----GGGGAUGGAGCCAUAAAAAUAAACCACCCAA--AAGUGGGCUAAAAUGAAGUUGUGUGUGCCAAGAACAAUAAAAAUGGCAAAGGAA--------- -----------((.(((((-----.(((.(((...............)))))).)--))))...))................(((((.............)))))......--------- ( -15.58) >DroVir_CAF1 28246 106 - 1 -----------GGCGGUGCGGGCAUGAUAUGGAGCCAUAAAAAUAAACCACCCAA-AAAGUGGGCUAAAAUGAAGUUCU--GUGCAAAGAACAAUAAAAAUGGCAGGCAGAGGACGCACG -----------....((((.(((....((((....))))........((((....-...)))))))........(((((--.(((......((.......))....))).))))))))). ( -27.00) >DroPse_CAF1 91589 105 - 1 GCUGGGCGGGGGCUCCUCC-----AGGGAUGGAGCCAUAAAAAUAAACCACCCAA-AAAGUGGGCUAAAAUGAAGUUGUGUGUGCGAAGAACAAUAAAAAUGGUAAAGGCA--------- (((.......((((((((.-----...)).))))))..........((((((((.-....))))..........(((((...........))))).....))))...))).--------- ( -27.30) >DroGri_CAF1 28559 94 - 1 -----------GUAAGGG-CGUCAUGAUAUGGAGCCAUACAAAUAAGCCACCCAAAAAAGUGGGCUAAAAUGAAGUUGU--GUGCAAAGAACAAUAAAAAUGGCAGGC------------ -----------(((..((-(..(((...)))..))).)))......((((((((......))))..........(((((--.........))))).....))))....------------ ( -20.80) >DroMoj_CAF1 23468 96 - 1 -----------GGCAGAG-GGCCCUGAUAUGGAGCCAUAAAAAUAAACCACCCAA-AAAGUGGGCUACAAUGAAGUUGU--GUGCAAAGAACAAUAAAAAUGGCAAGCCAG--------- -----------(((....-...((......)).(((((.........((((....-...))))((((((((...)))))--).))..............)))))..)))..--------- ( -24.00) >DroAna_CAF1 5076 104 - 1 GCUCGCC-GGGACAAAUUC-----AGGGAUGGAGCCAUAAAAAUAAACCACCCAA-AAAGUGGGCUAAAAUGAAGUUGUGUGUGCCAAGAACAAUAAAAAUGGCAAAGGAA--------- ((((((.-.(((....)))-----.(((.(((...............))))))..-...)))))).................(((((.............)))))......--------- ( -18.88) >consensus ___________GGAAGUGC_____UGAGAUGGAGCCAUAAAAAUAAACCACCCAA_AAAGUGGGCUAAAAUGAAGUUGU__GUGCAAAGAACAAUAAAAAUGGCAAACGAA_________ .................................(((((.........((((........))))...........(((((...........)))))....)))))................ (-12.52 = -12.55 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:24 2006