
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,790,796 – 10,790,912 |
| Length | 116 |
| Max. P | 0.928730 |

| Location | 10,790,796 – 10,790,912 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -23.87 |
| Energy contribution | -23.98 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
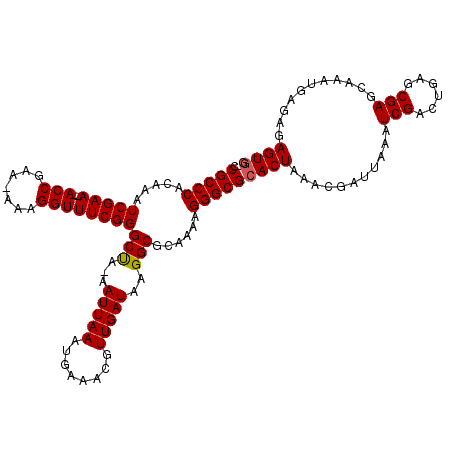
>3L_DroMel_CAF1 10790796 116 + 23771897 ACCGCCCACAAAUCGAA--ACCGAA-AAAGGUUUCGGGCUA-AAUUAAAUGAAACGUUGAUAAGGCGCAAAAGGGCGCACUAAACGAUUAAAUCGACUGAGCGAGCAAAUGAGAGAGUGG ..(((((.....(((((--(((...-...))))))))(((.-.(((((........)))))..)))......)))))((((...........(((......)))...........)))). ( -30.85) >DroYak_CAF1 143870 115 + 1 -GCGCCCACAAAUCGAA--ACCGAA-AAAGGUUUCGGGCUA-AAUUAAAUGAAACGUUGAUAAGGCGCAAAAGGGCGCACUAAACGAUUAAAUCGACUGAGCGAGCAAAUGAGAGAGUGG -((((((.....(((((--(((...-...))))))))(((.-.(((((........)))))..)))......))))))(((...........(((......)))...........))).. ( -32.15) >DroAna_CAF1 125779 118 + 1 GCCGCCCACAAAUCGAAAAACCGAAAAAAGGUUUCGGGCCAAAAUUAAAUGAAACGUUGAUAAGGCACAAAAGGGCGCACUAAACGGUUAAAUCGAAUGAGCGAGCAAAUGAGAGAGU-- (((((((.............((((((.....))))))(((...(((((........)))))..)))......))))).......((.(((.......))).)).))............-- ( -24.40) >consensus _CCGCCCACAAAUCGAA__ACCGAA_AAAGGUUUCGGGCUA_AAUUAAAUGAAACGUUGAUAAGGCGCAAAAGGGCGCACUAAACGAUUAAAUCGACUGAGCGAGCAAAUGAGAGAGUGG ..(((((.....(((((..(((.......))))))))(((...(((((........)))))..)))......)))))((((...........(((......)))...........)))). (-23.87 = -23.98 + 0.11)

| Location | 10,790,796 – 10,790,912 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.98 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.37 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10790796 116 - 23771897 CCACUCUCUCAUUUGCUCGCUCAGUCGAUUUAAUCGUUUAGUGCGCCCUUUUGCGCCUUAUCAACGUUUCAUUUAAUU-UAGCCCGAAACCUUU-UUCGGU--UUCGAUUUGUGGGCGGU ............((((((((..((((((......((((.((.((((......))))))....))))............-....((((((....)-))))).--.)))))).)))))))). ( -29.30) >DroYak_CAF1 143870 115 - 1 CCACUCUCUCAUUUGCUCGCUCAGUCGAUUUAAUCGUUUAGUGCGCCCUUUUGCGCCUUAUCAACGUUUCAUUUAAUU-UAGCCCGAAACCUUU-UUCGGU--UUCGAUUUGUGGGCGC- ..............((((((..((((((......((((.((.((((......))))))....))))............-....((((((....)-))))).--.)))))).))))))..- ( -28.80) >DroAna_CAF1 125779 118 - 1 --ACUCUCUCAUUUGCUCGCUCAUUCGAUUUAACCGUUUAGUGCGCCCUUUUGUGCCUUAUCAACGUUUCAUUUAAUUUUGGCCCGAAACCUUUUUUCGGUUUUUCGAUUUGUGGGCGGC --............((.(((((((((((...(((((...((.((((......))))))......((..(((........)))..))...........)))))..))))...))))))))) ( -28.00) >consensus CCACUCUCUCAUUUGCUCGCUCAGUCGAUUUAAUCGUUUAGUGCGCCCUUUUGCGCCUUAUCAACGUUUCAUUUAAUU_UAGCCCGAAACCUUU_UUCGGU__UUCGAUUUGUGGGCGG_ .............(((((((..((((((......((((.((.((((......))))))....)))).................(((((.......)))))....)))))).))))))).. (-26.25 = -26.37 + 0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:14 2006