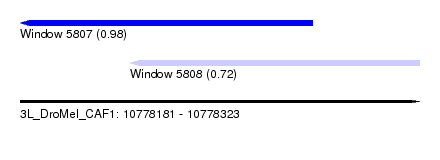
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,778,181 – 10,778,323 |
| Length | 142 |
| Max. P | 0.975148 |

| Location | 10,778,181 – 10,778,285 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -18.25 |
| Energy contribution | -17.69 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10778181 104 - 23771897 UCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUUCAUCGCAAAAAGGAUGUGAUGGAUGGAGCAUUUGUUCAAAUGCUAUUAGGACUCAAU-GAAAAGGAUAU ......((((...((((......((((((((.(((((((((((.......)))))))))))..).)))))))....)))).))))........-........... ( -24.40) >DroSec_CAF1 111779 105 - 1 UCAAAACUAAGCAGGCGGACCAUAACAAAUCACGUUCAUCGCAAAAAGGAUGUGAUGGAUGGAGCAUUUGUUCAAAUGCUAUUAGGACUCCUUCAAAAAGGAUAU ......((((...((((......((((((((.(((((((((((.......)))))))))))..).)))))))....)))).))))...(((((....)))))... ( -27.20) >DroEre_CAF1 117128 104 - 1 UCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUCCAUUGCGAAAAGGAUGUGAUGGAUGGAACAUUUUCUCAAAUGUUAUUAGGACGCCAU-AAGAAGGAUAU .............(((...((((((((.(((((((((.((....)).)))))))))...(((((....))).))..))))))).)...)))..-........... ( -23.00) >DroYak_CAF1 130243 86 - 1 UCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUCCAUCGCAAAAAGGAUGUGAUGGAAGGAAC--------AAAUGUUAUUAU-----------GAAGUAUAU ..........((...((...(((((((..((...(((((((((.......)))))))))..))..--------...))))))).)-----------)..)).... ( -17.60) >consensus UCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUCCAUCGCAAAAAGGAUGUGAUGGAUGGAACAUUUGUUCAAAUGCUAUUAGGACUCCAU_AAAAAGGAUAU ......((((...((((............((.(((((((((((.......))))))))))))).............)))).)))).................... (-18.25 = -17.69 + -0.56)


| Location | 10,778,220 – 10,778,323 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -20.81 |
| Consensus MFE | -15.98 |
| Energy contribution | -15.72 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10778220 103 - 23771897 -UCAAACCACCCAUAUCAACUGGGUAAAUCAAAGCAAUUUCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUUCAUCGCAAAAAGGAUGUGAUGGAUGGAGC -.....(((((((.......)))))........((..............)).))..............((.(((((((((((.......))))))))))))).. ( -21.34) >DroSec_CAF1 111819 101 - 1 -GC--CCCACCCAUAUCAACUGGUUAAAUCAAAGCAAUUUCAAAACUAAGCAGGCGGACCAUAACAAAUCACGUUCAUCGCAAAAAGGAUGUGAUGGAUGGAGC -((--.(((.((...((..(((.(((....(((....)))......))).)))...)).........(((((((((..........))))))))))).))).)) ( -20.60) >DroEre_CAF1 117167 102 - 1 UCC--CCCAACCAUAUCAGUUGAGUAAAUCAAAGCAAUUUCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUCCAUUGCGAAAAGGAUGUGAUGGAUGGAAC ...--.(((.((......(((.(((.....(((....)))....))).)))................(((((((((.((....)).))))))))))).)))... ( -20.60) >DroYak_CAF1 130264 102 - 1 AUC--CCCAACCAAAUCGGUUGAGUAAAUCAAAGCAAUUUCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUCCAUCGCAAAAAGGAUGUGAUGGAAGGAAC .((--(.(((((.....)))))...................................................(((((((((.......))))))))).))).. ( -20.70) >consensus _UC__CCCAACCAUAUCAACUGAGUAAAUCAAAGCAAUUUCAAAACUAAGCAGGCAGACAAUAACAAAUCACGUCCAUCGCAAAAAGGAUGUGAUGGAUGGAAC ......(((.((........(((.....)))....................................(((((((((..........))))))))))).)))... (-15.98 = -15.72 + -0.25)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:05:04 2006