
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,774,989 – 10,775,118 |
| Length | 129 |
| Max. P | 0.976630 |

| Location | 10,774,989 – 10,775,092 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.47 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10774989 103 - 23771897 --------AUUCGAGGCAAUA---CGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----GUGCACUUCGAACACCCCCGGGCGAGUUAAAAGCAUUAACACAGGCU--CGAGUGGCU --------(((((((.(....---.....((((((((..(((((((....))))))).----(((.........)))..))))))))(((((.....)))))...).))--))))).... ( -42.50) >DroSec_CAF1 108563 103 - 1 --------AUUCGAGGCACCA---CGGCCUUGUGCGGAAGCACCACGCAAGUGGUGCA----GUGCACUUCGAACACCCCCGGGCGAGUUAACAGCAUUAACACAGGCU--CGAGUGGCU --------.((((((((....---..)))..(((((...(((((((....))))))).----.))))).)))))(((...(((((..(((((.....)))))....)))--)).)))... ( -40.20) >DroSim_CAF1 111520 103 - 1 --------AUUCGAGGCACCA---CGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----GUGCACUUCGAACACCCCCGGGCGAGUUAACAGCAUUAACACAGGCU--CGAGUGGCU --------.....((.(((..---(((((((((((((..(((((((....))))))).----(((.........)))..))))))))(((((.....)))))...))).--)).))).)) ( -43.10) >DroEre_CAF1 113842 114 - 1 CCACCGCCAUCCGAGGCACCACCGCGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----CUGCACUUCGAACACCCCCGGGCCAGUUAACAGUAUUAACACAGGCU--CGAGUGGCU .....(((......))).((((((.(((((.((((((..(((((((....))))))).----(........).......))))))..(((((.....)))))..)))))--)).)))).. ( -45.60) >DroYak_CAF1 125185 106 - 1 --------AUUCGCGGCUCCACCGCGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----CUGCACUUCGAACACCCCCGGGCCAGUUAACAGCAUUAACACAGGCU--CGAGUGGCU --------...(((((.....)))))(((..((.(((..(((((((....))))))).----))).))............((((((.(((((.....)))))...))))--))...))). ( -42.50) >DroAna_CAF1 109295 103 - 1 --------AUUCGAG-----A---CGGCCUUGUGCGGAAGC-CAACGCAAGUGGUGCAAGUGAUGCACUACGACACCCCCGGGGCAAGUUGGCCAAGUUAACACAGGCUCUCGAGAGCCU --------.((((((-----(---..((((.(((..((.((-((((((..((((((((.....))))))))....((....))))..))))))....))..))))))))))))))..... ( -42.30) >consensus ________AUUCGAGGCACCA___CGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA____GUGCACUUCGAACACCCCCGGGCGAGUUAACAGCAUUAACACAGGCU__CGAGUGGCU ..............(((........(((((.((((((..(((((((....)))))))........(.....).......))))))..(((((.....)))))..)))))........))) (-30.03 = -30.92 + 0.89)

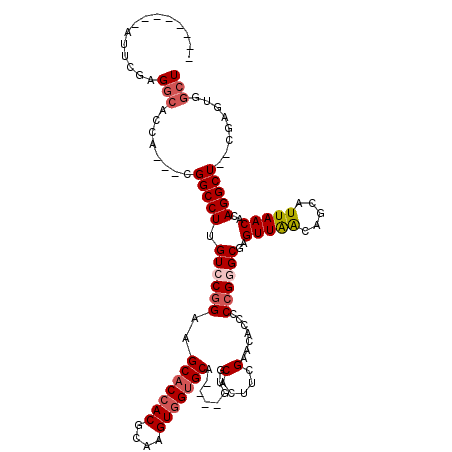
| Location | 10,775,027 – 10,775,118 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Mean single sequence MFE | -35.82 |
| Consensus MFE | -19.73 |
| Energy contribution | -20.57 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10775027 91 - 23771897 ACACUUUUAU-AGAG--------CGCACCACCG---CG----------AUUCGAGGCAAUA---CGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----GUGCACUUCGAACACCCC ..........-.(((--------.((((..(((---.(----------(..((((((....---..)))))))))))..(((((((....))))))).----)))).))).......... ( -38.00) >DroSec_CAF1 108601 91 - 1 ACACUUUUAU-AGAG--------CGCACCACCG---CG----------AUUCGAGGCACCA---CGGCCUUGUGCGGAAGCACCACGCAAGUGGUGCA----GUGCACUUCGAACACCCC ..........-.(((--------.((((..(((---((----------...((((((....---..)))))))))))..(((((((....))))))).----)))).))).......... ( -40.50) >DroSim_CAF1 111558 92 - 1 ACACUUUUAUGAGAG--------CGCACCACCG---CG----------AUUCGAGGCACCA---CGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----GUGCACUUCGAACACCCC .........(((.((--------.((((..(((---.(----------(..((((((....---..)))))))))))..(((((((....))))))).----)))).)))))........ ( -38.30) >DroEre_CAF1 113880 104 - 1 ACACUUUUAU-AGAG--------CGAACCACCG---CGAACCACCGCCAUCCGAGGCACCACCGCGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----CUGCACUUCGAACACCCC ..........-.(..--------((((...(((---((.......(((......))).....)))))....((.(((..(((((((....))))))).----))).))))))..)..... ( -32.90) >DroYak_CAF1 125223 94 - 1 ACACUUUUAU-AGAG--------CGAACCACCG---CG----------AUUCGCGGCUCCACCGCGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA----CUGCACUUCGAACACCCC ..........-.(..--------((((...(((---.(----------((((((((.....))))))....))))))..(((((((....))))))).----......))))..)..... ( -34.30) >DroAna_CAF1 109335 100 - 1 GCAUUUAUAU-AGCUGUAUGUUUCCAGUUGCGGUGAUG----------AUUCGAG-----A---CGGCCUUGUGCGGAAGC-CAACGCAAGUGGUGCAAGUGAUGCACUACGACACCCCC .........(-(((((........)))))).((((...----------.......-----.---..((.(((.((....))-))).))..((((((((.....))))))))..))))... ( -30.90) >consensus ACACUUUUAU_AGAG________CGAACCACCG___CG__________AUUCGAGGCACCA___CGGCCUUGUCCGGAAGCACCACGCAAGUGGUGCA____GUGCACUUCGAACACCCC ...................................................((((((.........)))))).......(((((((....)))))))....................... (-19.73 = -20.57 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:57 2006