
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,772,763 – 10,772,876 |
| Length | 113 |
| Max. P | 0.939082 |

| Location | 10,772,763 – 10,772,876 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.45 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -23.75 |
| Energy contribution | -24.42 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10772763 113 + 23771897 CGCAUUGAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGCAAAAAUUUCAGGCAGGUU--GACUGACAAAAAAAAAAGAAAAA-AAAA----UGA ((((((......((((((.......)))))).....))))))((((((((((((((............)))))).)))).)))--)......................-....----... ( -27.30) >DroSec_CAF1 106331 105 + 1 CGCAUUGAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGCAAAAAUUUCAGGCAGGUU--GACUGACAAAAAAA--------A-AAAA----UGU ((((((......((((((.......)))))).....))))))((((((((((((((............)))))).)))).)))--).............--------.-....----... ( -27.30) >DroSim_CAF1 109293 102 + 1 CGCAUUGAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGCAAAAAUUUCAGGCAGGUU--GACUGACAAAA-----------A-AAAA----UGG ((((((......((((((.......)))))).....))))))((((((((((((((............)))))).)))).)))--)..........-----------.-....----... ( -27.30) >DroEre_CAF1 111481 111 + 1 CGCAUUGAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGUAAAAAUUUCAGGCAGGUU--GACUGACAAAAAAAG---AAAAAAUACG----GAG ((((((......((((((.......)))))).....))))))((.(((((((((((............)))))).)))))(((--....)))........---........))----... ( -27.90) >DroYak_CAF1 122757 105 + 1 CGCAUUAAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGUAAAAAUUUCAGGCAGGUU--GACUGACAAAAUAAA---------GUGG----UGG ((((((......((((((.......)))))).....))))))((((((((((((((............)))))).)))).)))--)(((.((........---------))))----).. ( -26.40) >DroAna_CAF1 107283 117 + 1 CGCAUUGAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGAAAAAAUAUCAACCAAGUAGCGACAGCCCCAAAAAA---AAACUUGACAGCCCUGG ((((((......((((((.......)))))).....))))))...(((((((.....(((((((......(((.......)))((....)).))))))).---....)))))))...... ( -31.20) >consensus CGCAUUGAAAAUCGCUGCAAAUGAGGCAGCGUGAAAAAUGCGCGACUGUCGAAAUUGUUUUUGGCAAAAAUUUCAGGCAGGUU__GACUGACAAAAAAAA_______A_AAAA____UGG ((((((......((((((.......)))))).....))))))...(((((((((((............)))))).)))))........................................ (-23.75 = -24.42 + 0.67)

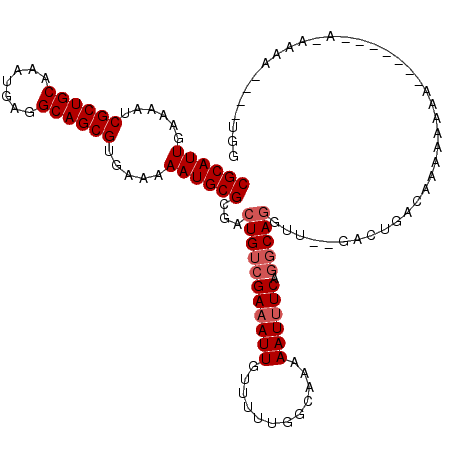

| Location | 10,772,763 – 10,772,876 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.45 |
| Mean single sequence MFE | -23.35 |
| Consensus MFE | -18.56 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682841 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10772763 113 - 23771897 UCA----UUUU-UUUUUCUUUUUUUUUUGUCAGUC--AACCUGCCUGAAAUUUUUGCCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUCAAUGCG ...----....-..............(((((((.(--.....).))((((((.((......)).)))))))))))...((((((.....((((((.......))))))......)))))) ( -21.10) >DroSec_CAF1 106331 105 - 1 ACA----UUUU-U--------UUUUUUUGUCAGUC--AACCUGCCUGAAAUUUUUGCCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUCAAUGCG ...----....-.--------.....(((((((.(--.....).))((((((.((......)).)))))))))))...((((((.....((((((.......))))))......)))))) ( -21.10) >DroSim_CAF1 109293 102 - 1 CCA----UUUU-U-----------UUUUGUCAGUC--AACCUGCCUGAAAUUUUUGCCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUCAAUGCG ...----....-.-----------..(((((((.(--.....).))((((((.((......)).)))))))))))...((((((.....((((((.......))))))......)))))) ( -21.10) >DroEre_CAF1 111481 111 - 1 CUC----CGUAUUUUUU---CUUUUUUUGUCAGUC--AACCUGCCUGAAAUUUUUACCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUCAAUGCG ...----..........---......(((((((.(--.....).))((((((.((......)).)))))))))))...((((((.....((((((.......))))))......)))))) ( -20.20) >DroYak_CAF1 122757 105 - 1 CCA----CCAC---------UUUAUUUUGUCAGUC--AACCUGCCUGAAAUUUUUACCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUUAAUGCG ...----....---------......(((((((.(--.....).))((((((.((......)).)))))))))))...((((((.....((((((.......))))))......)))))) ( -20.90) >DroAna_CAF1 107283 117 - 1 CCAGGGCUGUCAAGUUU---UUUUUUGGGGCUGUCGCUACUUGGUUGAUAUUUUUUCCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUCAAUGCG ....((((((((((((.---.(((((((((.(((((((....)).)))))....))))))))).))))).))))))).((((((.....((((((.......))))))......)))))) ( -35.70) >consensus CCA____UUUU_U_______UUUUUUUUGUCAGUC__AACCUGCCUGAAAUUUUUGCCAAAAACAAUUUCGACAGUCGCGCAUUUUUCACGCUGCCUCAUUUGCAGCGAUUUUCAAUGCG ........................................(((..(((((((.((......)).))))))).)))...((((((.....((((((.......))))))......)))))) (-18.56 = -18.58 + 0.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:53 2006