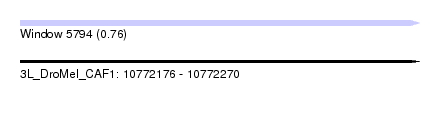
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,772,176 – 10,772,270 |
| Length | 94 |
| Max. P | 0.763237 |

| Location | 10,772,176 – 10,772,270 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.39 |
| Mean single sequence MFE | -34.39 |
| Consensus MFE | -20.97 |
| Energy contribution | -20.37 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
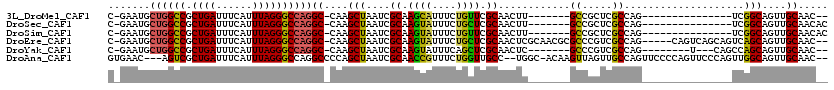
>3L_DroMel_CAF1 10772176 94 + 23771897 C-GAAUGCUGGCCGCUGAUUUCAUUUAGGGCCAGGC-CAAGCUAAUCGCAAGCAUUUCUGUUCGCAACUU-------GCCGCUCGCCAG---------------UCGGCAGUUGCAAC-- .-((((.((((((.((((......))))))))))..-...(((.......)))......))))((((((.-------((((((....))---------------.))))))))))...-- ( -36.10) >DroSec_CAF1 105788 96 + 1 C-GAAUGCUGGCCGCUGAUUUCAUUUAGGGCCAGGC-CAAGCUAAUCGCAAGUAUUUCUGCUCGCAACUU-------GCCGCUCGCCAG---------------UCGGCAGUUGCAACAC .-.....((((((.((((......))))))))))..-..(((.((((....).)))...))).((((((.-------((((((....))---------------.))))))))))..... ( -35.20) >DroSim_CAF1 108693 96 + 1 C-GAAUGCUGGCCGCUGAUUUCAUUUAGGGCCAGGC-CAAGCUAAUCGCAAGUAUUUCUGUUCGCAACUU-------GCCGCUCGCCAG---------------UCGGCAGUUGCAACAC .-((((.((((((.((((......))))))))))..-...((.....))..........))))((((((.-------((((((....))---------------.))))))))))..... ( -35.20) >DroEre_CAF1 110792 111 + 1 C-GAAUGCUGGCCGCUGAUUUCAUUUAGGGCCAGGC-CAAGCUAAUCGCAAGUAUUUCUGCUCGCAACUCGCAACGCGCCCGUCGCCAG-----CAGUCAGCAGUCAGCAGUUGCAAC-- (-((.(((((((.(((((((........(((...))-)..(((....((.((((....)))).)).....((.(((....))).)).))-----)))))))).))))))).)))....-- ( -35.70) >DroYak_CAF1 122103 98 + 1 C-GAAUGCUGGCCGCUGAUUUCAUUUAGGGCCAGGC-CAAGCUAAUCGCAAGUAUUUCAGCUCGCAACUC-------GCCCGUCGCCAG--------U---CAGCCAGCAGUUGCAAC-- (-((.(((((((...(((((.......((((.((..-(.((((((((....).)))..)))).)...)).-------))))......))--------)---))))))))).)))....-- ( -30.42) >DroAna_CAF1 106623 112 + 1 GUGAAC---AGUCGCUGAUUUCAUUUAGGGCCAGGCCCCAGCUAAUCGCAACCGUUUCUGGUUGCC--UGGC-ACAAGUUAGUUGCCAGUUCCCCAGUUCCCAGUUGGCAGUUGCAAC-- ......---.((.((((..........((((...))))(((((....(((((((....)))))))(--((((-((......).)))))).............))))).)))).))...-- ( -33.70) >consensus C_GAAUGCUGGCCGCUGAUUUCAUUUAGGGCCAGGC_CAAGCUAAUCGCAAGUAUUUCUGCUCGCAACUU_______GCCGCUCGCCAG_______________UCGGCAGUUGCAAC__ .......((((((.((((......))))))))))((....(((....((.((((....)))).))............((.....))....................)))....))..... (-20.97 = -20.37 + -0.61)
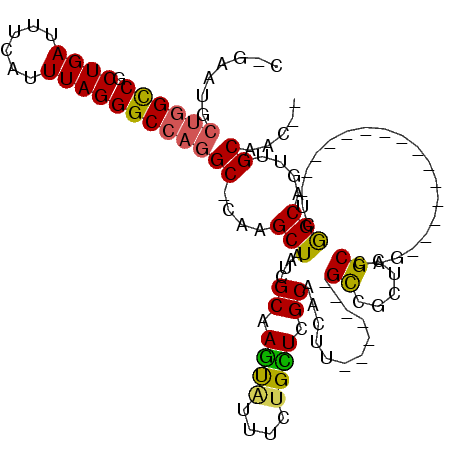

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:51 2006