
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,771,894 – 10,772,035 |
| Length | 141 |
| Max. P | 0.999904 |

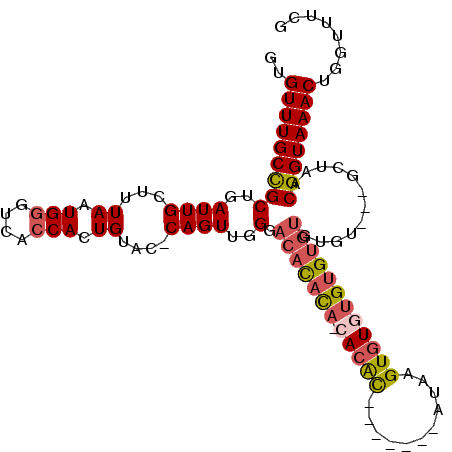
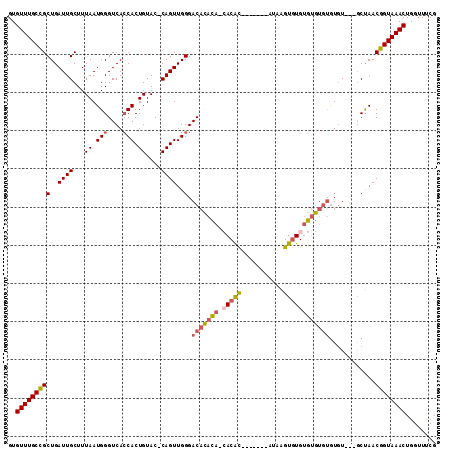
| Location | 10,771,894 – 10,771,995 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -21.00 |
| Energy contribution | -22.24 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10771894 101 + 23771897 GUGUUUGCCGCUGAUUGCUUUAAUGGGUCAGCACUGUAC-CAGUUGGGACAUAC---ACAC-------AGAAGUGUGUGUGUGUGUGUUGCACUAACGGUAAACUGGUUUCG ..((((((((((((((.(......)))))))).......-..((((((((((((---((((-------(.(....).))))))))))))...))))))))))))........ ( -38.00) >DroSec_CAF1 105530 90 + 1 GUGUUUGCCGCUGAUUGCUUUAAUGGGUCACCACUGUAC-CAGUUGGGAC-------ACAC-------AAAAGUGUG----UGUGUGU---GCUAACGGUAAACUGGUUUCG ..(((((((((.....)).......(((((....)).))-).(((((.((-------((((-------(........----)))))))---.))))))))))))........ ( -29.50) >DroSim_CAF1 108425 100 + 1 GUGUUUGCCGCUGAUUGCUUUAAUGGGUCACCACUGUAC-CAGUUGGGACAUACA-CACAC-------AUAAGUGUGUGUGUGUGUGU---GCUAACGGUAAACUGGUUUCG ..(((((((((.....)).......(((((....)).))-).(((((.(((((((-(((((-------((....))))))))))))))---.))))))))))))........ ( -42.10) >DroEre_CAF1 110509 106 + 1 GUGUUUGCCGCUGAUUGCUUUAAUGGGUCACCACUGUAC-CAGUUGGGACACAUA-CACGUAUA-UAUGCAAGUGUGUGUGUGUGUGU---GCUAACGGUAAACUGGUUUCG ..(((((((((.....)).......(((((....)).))-).(((((.(((((((-((((((((-(......))))))))))))))))---.))))))))))))........ ( -39.40) >DroAna_CAF1 106382 106 + 1 GUGUUUGCUGCUGAUUGCUUUAAUGGGUCACCACUGUACCCAGUUGGGACACCCAACACACAUACU--AUAGGUAUGUGCGU-CAUUU---GCCAACGGUAAACUGGUUUCG ..((((((((.((...((.....(((((..((((((....))).)))...)))))((.(((((((.--....))))))).))-.....---)))).))))))))........ ( -32.70) >consensus GUGUUUGCCGCUGAUUGCUUUAAUGGGUCACCACUGUAC_CAGUUGGGACACACA_CACAC_______AUAAGUGUGUGUGUGUGUGU___GCUAACGGUAAACUGGUUUCG ..(((((((((..((((...((.(((....))).))....))))..).(((((((.(((((...........))))))))))))............))))))))........ (-21.00 = -22.24 + 1.24)

| Location | 10,771,894 – 10,771,995 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.66 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.796149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10771894 101 - 23771897 CGAAACCAGUUUACCGUUAGUGCAACACACACACACACACUUCU-------GUGU---GUAUGUCCCAACUG-GUACAGUGCUGACCCAUUAAAGCAAUCAGCGGCAAACAC ........((((.(((((((..(.......(((((((......)-------))))---)).(((.((....)-).))))..)))))........((.....)))).)))).. ( -28.10) >DroSec_CAF1 105530 90 - 1 CGAAACCAGUUUACCGUUAGC---ACACACA----CACACUUUU-------GUGU-------GUCCCAACUG-GUACAGUGGUGACCCAUUAAAGCAAUCAGCGGCAAACAC ........((((.((((((.(---(((((((----((......)-------))))-------)).((....)-)....))).))))........((.....)))).)))).. ( -22.90) >DroSim_CAF1 108425 100 - 1 CGAAACCAGUUUACCGUUAGC---ACACACACACACACACUUAU-------GUGUG-UGUAUGUCCCAACUG-GUACAGUGGUGACCCAUUAAAGCAAUCAGCGGCAAACAC ........((((.((((((.(---((..(((((((((......)-------)))))-))).(((.((....)-).)))))).))))........((.....)))).)))).. ( -30.60) >DroEre_CAF1 110509 106 - 1 CGAAACCAGUUUACCGUUAGC---ACACACACACACACACUUGCAUA-UAUACGUG-UAUGUGUCCCAACUG-GUACAGUGGUGACCCAUUAAAGCAAUCAGCGGCAAACAC ........((((.((((((.(---((....(((((.((((.......-.....)))-).))))).((....)-)....))).))))........((.....)))).)))).. ( -23.30) >DroAna_CAF1 106382 106 - 1 CGAAACCAGUUUACCGUUGGC---AAAUG-ACGCACAUACCUAU--AGUAUGUGUGUUGGGUGUCCCAACUGGGUACAGUGGUGACCCAUUAAAGCAAUCAGCAGCAAACAC ........((((.(.((((((---....(-((((((((((....--.))))))))))).((.((((((.(((....)))))).)))))......))...)))).).)))).. ( -34.70) >consensus CGAAACCAGUUUACCGUUAGC___ACACACACACACACACUUAU_______GUGUG_UGUAUGUCCCAACUG_GUACAGUGGUGACCCAUUAAAGCAAUCAGCGGCAAACAC ........((((.(((((.................(((((...........)))))......((((((.(((....)))))).)))..............))))).)))).. (-14.62 = -15.66 + 1.04)

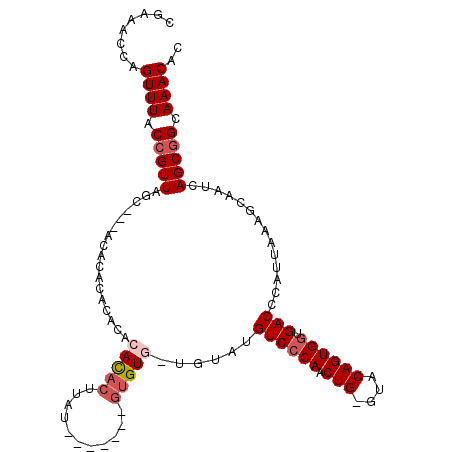
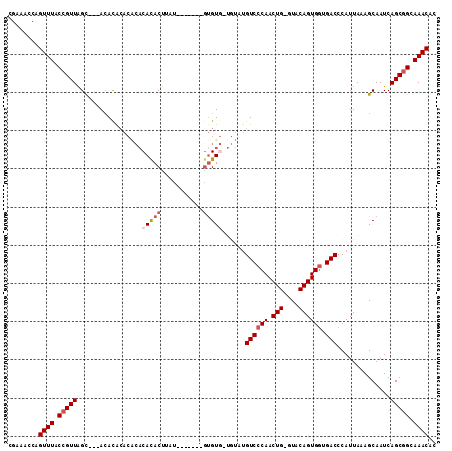
| Location | 10,771,933 – 10,772,035 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -13.34 |
| Energy contribution | -14.86 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.42 |
| SVM decision value | 4.47 |
| SVM RNA-class probability | 0.999904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
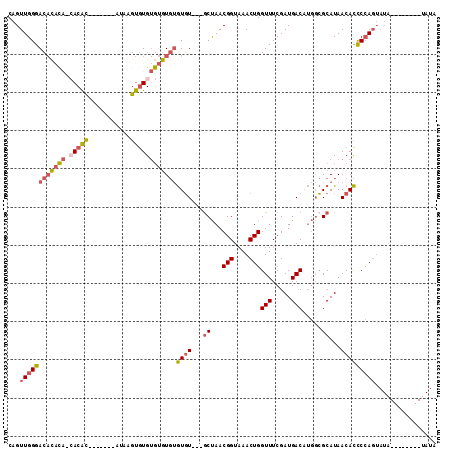
>3L_DroMel_CAF1 10771933 102 + 23771897 CAGUUGGGACAUAC---ACAC-------AGAAGUGUGUGUGUGUGUGUUGCACUAACGGUAAACUGGUUUCGAUGACAUGGCGCAUAACACCCCAGUAUAUAUAUAUAUAUA ....((((((((((---((((-------....))))))))))((((..(((.(((.(((....)))(((.....))).))).)))..))))))))((((((....)))))). ( -33.50) >DroSec_CAF1 105569 83 + 1 CAGUUGGGAC-------ACAC-------AAAAGUGUG----UGUGUGU---GCUAACGGUAAACUGGUUUCGAUGACAUGGCGCAUAACACCCCAGUAUA--------UAUA ...(((((((-------((((-------....)))))----).(((((---((((.(((....)))(((.....))).)))))))))....)))))....--------.... ( -27.10) >DroSim_CAF1 108464 93 + 1 CAGUUGGGACAUACA-CACAC-------AUAAGUGUGUGUGUGUGUGU---GCUAACGGUAAACUGGUUUCGAUGACAUGGCGCAUAACACCCCAGUAUA--------UAUA ...((((((((((((-(((((-------....))))))))))))((((---((((.(((....)))(((.....))).)))))))).....)))))....--------.... ( -37.00) >DroEre_CAF1 110548 99 + 1 CAGUUGGGACACAUA-CACGUAUA-UAUGCAAGUGUGUGUGUGUGUGU---GCUAACGGUAAACUGGUUUCGAUGACAUGGCGCAUAACACCCCAGUAUA--------UAUA ..(((((.(((((((-((((((((-(......))))))))))))))))---.))))).(((.(((((.....(((........)))......))))).))--------)... ( -34.10) >DroAna_CAF1 106422 91 + 1 CAGUUGGGACACCCAACACACAUACU--AUAGGUAUGUGCGU-CAUUU---GCCAACGGUAAACUGGUUUCGAUGACUACUAGCAUAACAUUCAA---------------CA ..((((((...)))))).(((((((.--....))))))).((-((((.---((((.........))))...))))))..................---------------.. ( -25.30) >consensus CAGUUGGGACACACA_CACAC_______AUAAGUGUGUGUGUGUGUGU___GCUAACGGUAAACUGGUUUCGAUGACAUGGCGCAUAACACCCCAGUAUA________UAUA ...((((((((((((.(((((...........))))))))))))((((...((...(((....)))(((.....))).....))...)))))))))................ (-13.34 = -14.86 + 1.52)


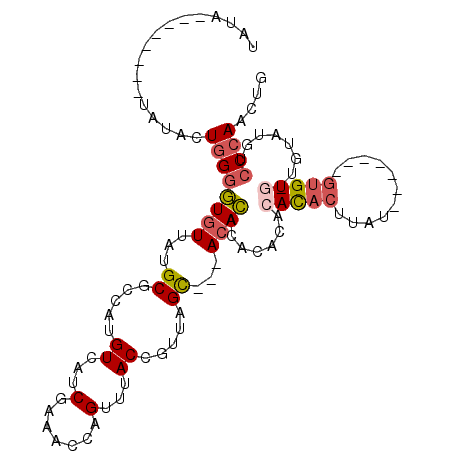
| Location | 10,771,933 – 10,772,035 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -6.54 |
| Energy contribution | -7.30 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.28 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
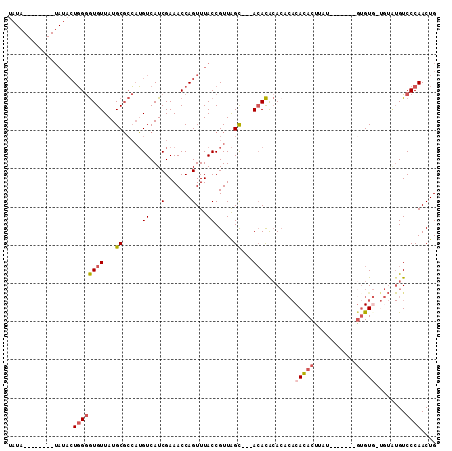
>3L_DroMel_CAF1 10771933 102 - 23771897 UAUAUAUAUAUAUAUACUGGGGUGUUAUGCGCCAUGUCAUCGAAACCAGUUUACCGUUAGUGCAACACACACACACACACUUCU-------GUGU---GUAUGUCCCAACUG ((((((....)))))).((((((((..(((((.(((....(.......).....)))..)))))..))))(((.((((((....-------))))---)).))))))).... ( -24.20) >DroSec_CAF1 105569 83 - 1 UAUA--------UAUACUGGGGUGUUAUGCGCCAUGUCAUCGAAACCAGUUUACCGUUAGC---ACACACA----CACACUUUU-------GUGU-------GUCCCAACUG ....--------((.(((((((((.....)))).((....))...))))).))..(((.(.---(((((((----........)-------))))-------)).).))).. ( -20.30) >DroSim_CAF1 108464 93 - 1 UAUA--------UAUACUGGGGUGUUAUGCGCCAUGUCAUCGAAACCAGUUUACCGUUAGC---ACACACACACACACACUUAU-------GUGUG-UGUAUGUCCCAACUG ....--------((.(((((((((.....)))).((....))...))))).))..(((.(.---(((.(((((((((......)-------)))))-))).))).).))).. ( -27.30) >DroEre_CAF1 110548 99 - 1 UAUA--------UAUACUGGGGUGUUAUGCGCCAUGUCAUCGAAACCAGUUUACCGUUAGC---ACACACACACACACACUUGCAUA-UAUACGUG-UAUGUGUCCCAACUG ....--------((.(((((((((.....)))).((....))...))))).))..(((.(.---(((((.((((.............-.....)))-).))))).).))).. ( -20.57) >DroAna_CAF1 106422 91 - 1 UG---------------UUGAAUGUUAUGCUAGUAGUCAUCGAAACCAGUUUACCGUUGGC---AAAUG-ACGCACAUACCUAU--AGUAUGUGUGUUGGGUGUCCCAACUG .(---------------(((((((.((.(((.((..........)).))).)).))))(((---(..((-((((((((((....--.))))))))))))..)))).)))).. ( -25.80) >consensus UAUA________UAUACUGGGGUGUUAUGCGCCAUGUCAUCGAAACCAGUUUACCGUUAGC___ACACACACACACACACUUAU_______GUGUG_UGUAUGUCCCAACUG .................((((((((...((.....((...(.......)...)).....))...)))).......(((((...........)))))........)))).... ( -6.54 = -7.30 + 0.76)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:50 2006