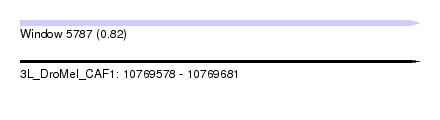
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,769,578 – 10,769,681 |
| Length | 103 |
| Max. P | 0.815421 |

| Location | 10,769,578 – 10,769,681 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.58 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -25.19 |
| Energy contribution | -25.58 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10769578 103 + 23771897 GCUCAUUUCGCACAAGACUUUCAAAUGGCUAAGACAC-AAAC-----UCAAAGUGAGGUAAAGCCAAAACG----U----UAACUUGGCCAUUGGCGAUCUUUGCAGUGAGCUUCGA--- (((((((..(((.((((......((((((((((.(((-....-----.....))).((.....))......----.----...)))))))))).....)))))))))))))).....--- ( -31.20) >DroSec_CAF1 103221 103 + 1 GCUCAUUUCGCACAAGACUUUCAAAUGGCUAAGACACCAAAC-----UCAAAGUGAGGUAAAGCCA-AACG----U----UAACUUGGCCAUUGGCGAUCUUUGCAGUGAGCUUCGA--- (((((((..(((.((((......((((((((((.(((.....-----.....))).((.....)).-....----.----...)))))))))).....)))))))))))))).....--- ( -30.60) >DroSim_CAF1 106115 103 + 1 GCUCAUUUCGCACAAGACUUUCAAAUGGCUAAGACACCAAAC-----UCAAAGUGAGGUAAAGCCA-AACG----U----UAACUUGGCCAUUGGCGAUCUUUGCAGUGAGCUUCGA--- (((((((..(((.((((......((((((((((.(((.....-----.....))).((.....)).-....----.----...)))))))))).....)))))))))))))).....--- ( -30.60) >DroEre_CAF1 108110 105 + 1 GCUCAUUUCGCACAAGACUUUCAAAUGGCUAAGACAC-AAAC-----UCAAAGUGAGGUAAAGCCA-AACG----U----UAACUUGGCCAUUGGCGAUCUUUGCAGUGAGCUUCGAGGU (((((((..(((.((((......((((((((((.(((-....-----.....))).((.....)).-....----.----...)))))))))).....))))))))))))))........ ( -31.20) >DroYak_CAF1 119410 105 + 1 GCUCAUUUCGCACAAGACUUUCAAAUGGCUAAGACAC-CAGC-----UCAAAGUGAGGUAAAGCCA-AACG----U----UAACUUGGCCAUUGGCGAUCUUUGCAGUGAGCUUCGAAAU (((((((..(((.((((......((((((((((.(((-....-----.....))).((.....)).-....----.----...)))))))))).....))))))))))))))........ ( -30.70) >DroAna_CAF1 104238 116 + 1 GCUCUUUUCGCACAAGACUUUCAAAUGGCUAAAAGCUCAAACUGAUCUCCAACUGAAGCAGAGCCA-GGCCAAAGCAAAAAGACUUGGCCAUUGGCGAUCUUUGCAGCGGACUCUGA--- ((.....((((.(((..........(((((....(((((...((.....))..)).)))..)))))-((((((..(.....)..)))))).))))))).....))............--- ( -29.60) >consensus GCUCAUUUCGCACAAGACUUUCAAAUGGCUAAGACAC_AAAC_____UCAAAGUGAGGUAAAGCCA_AACG____U____UAACUUGGCCAUUGGCGAUCUUUGCAGUGAGCUUCGA___ (((((((..(((.((((......((((((((((..............((.....))(((...)))..................)))))))))).....))))))))))))))........ (-25.19 = -25.58 + 0.39)

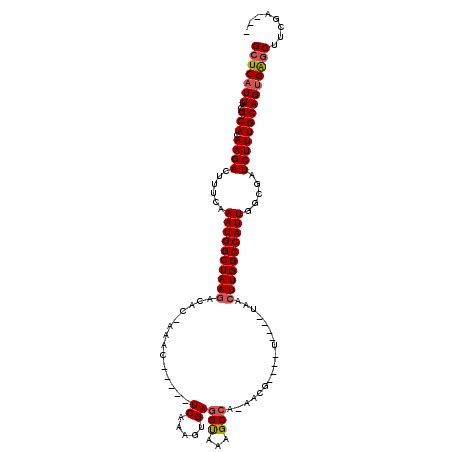
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:45 2006