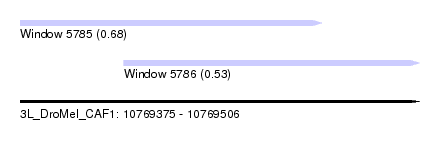
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,769,375 – 10,769,506 |
| Length | 131 |
| Max. P | 0.680257 |

| Location | 10,769,375 – 10,769,474 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -19.52 |
| Consensus MFE | -13.15 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
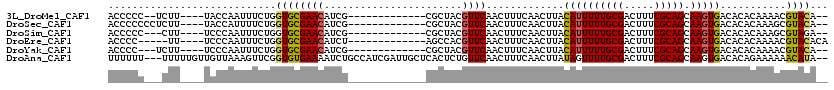
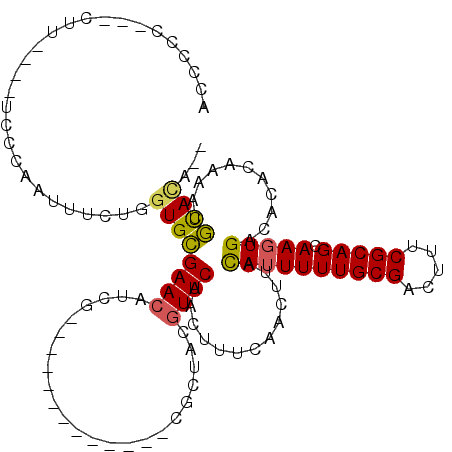
>3L_DroMel_CAF1 10769375 99 + 23771897 ACCCCC--UCUU----UACCAAUUUCUGGUGCGAACAUCG-------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACA-- ......--....----(((.......(((((((.....))-------------)))))..................((((((((((.....))))).)))))...........)))..-- ( -18.70) >DroSec_CAF1 103022 101 + 1 ACCCCCCCUCUU----UACCAUUUUCUGGUGCGAACAUCG-------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAGCGUACA-- .......(.(((----(.........(((((((.....))-------------)))))..................((((((((((.....))))).)))))......)))).)....-- ( -19.20) >DroSim_CAF1 105918 98 + 1 ACCCCC---CUU----UCCCAAUUUCUGGUGCGAACAUCG-------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAGCGUAGA-- .(..(.---(((----(.........(((((((.....))-------------)))))..................((((((((((.....))))).)))))......)))).)..).-- ( -18.80) >DroEre_CAF1 107899 98 + 1 ACCCC-----UU----UCCCAAUUUCUGGUGCGAACAUCU-------------AGCCACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACACA .....-----..----............((((((((....-------------......)))).............((((((((((.....))))).)))))...........))))... ( -16.40) >DroYak_CAF1 119210 98 + 1 ACCCC---UCUU----UCCCAAUUUCUGGUGCGAACAUCG-------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACA-- .....---....----..........(((((((.....))-------------)))))..................((((((((((.....))))).)))))................-- ( -18.50) >DroAna_CAF1 104044 115 + 1 UUUUUU---UUUUUGUUGUUAAAGUUCGGUGUGAAAAUCUGCCAUCGAUUGCUCACUCUGUUCAACUUUCAACUUAUAGUUUUGCGACUUUCGCAGCAAGUGACACAGAAAAAACAUA-- ...(((---(((((((.((((..(..(((.((((.((((.......))))..)))).)))..)..................(((((.....)))))....))))))))))))))....-- ( -25.50) >consensus ACCCCC___CUU____UCCCAAUUUCUGGUGCGAACAUCG_____________CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACA__ ............................((((((((.......................)))).............((((((((((.....))))).)))))...........))))... (-13.15 = -12.98 + -0.16)

| Location | 10,769,409 – 10,769,506 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.86 |
| Mean single sequence MFE | -13.30 |
| Consensus MFE | -10.70 |
| Energy contribution | -11.14 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.87 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
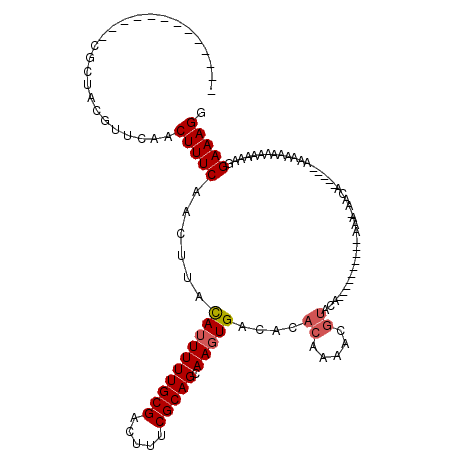
>3L_DroMel_CAF1 10769409 97 + 23771897 -------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACA----------AAACAACAAAACAAAAAAAAAAAAAGGAAAGG -------------............(((((......((((((((((.....))))).)))))...........((...----------..))......................))))). ( -13.40) >DroSec_CAF1 103058 88 + 1 -------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAGCGUACA----------AAAAAAUA--------AAAA-AAAAGGAAAGG -------------............(((((......((((((((((.....))))).)))))((.(.....).))...----------........--------....-.....))))). ( -13.40) >DroSim_CAF1 105951 89 + 1 -------------CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAGCGUAGA----------AAAAAAUA--------AAAAAAAAAGGAAAGG -------------..(((((((...................(((((.....)))))...(((...)))..))))))).----------........--------................ ( -13.40) >DroEre_CAF1 107930 104 + 1 -------------AGCCACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACACAGAUAUAUAAAA-AACAAAA--AAAAAAACAAAAGGAAAGG -------------............(((((......((((((((((.....))))).)))))....((.....))................-.......--.............))))). ( -12.90) >DroAna_CAF1 104081 100 + 1 GCCAUCGAUUGCUCACUCUGUUCAACUUUCAACUUAUAGUUUUGCGACUUUCGCAGCAAGUGACACAGAAAAAACAUA----------GAA----------AAAUAACAAAAAGGAAAGG .((...((....))..((((((((.(((.((((.....))).((((.....))))).)))))).))))).........----------...----------............))..... ( -13.40) >consensus _____________CGCUACGUUCAACUUUCAACUUACAUUUUUGCGACUUUCGCAGCAAGUGACACACAAAACGUACA__________AAA_AACA_____AAAAAAAAAAAAGGAAAGG .........................(((((......((((((((((.....))))).)))))....((.....)).......................................))))). (-10.70 = -11.14 + 0.44)

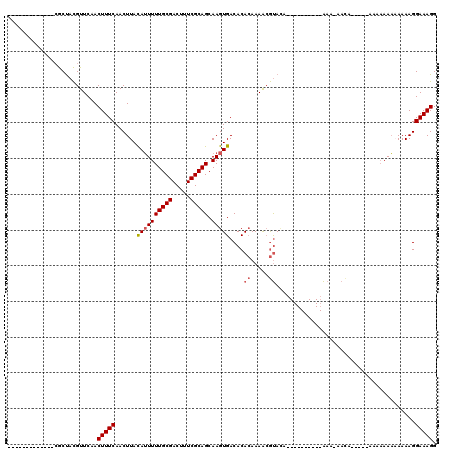
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:44 2006