
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,768,116 – 10,768,212 |
| Length | 96 |
| Max. P | 0.975652 |

| Location | 10,768,116 – 10,768,212 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.83 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.57 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
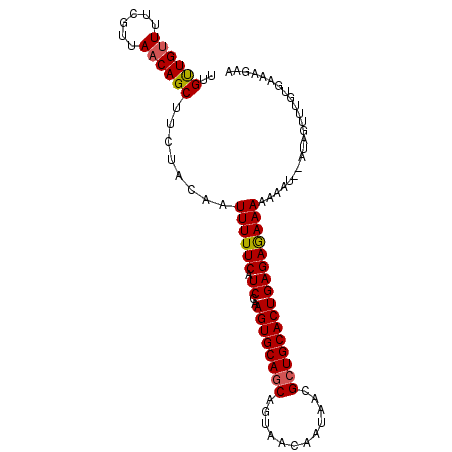
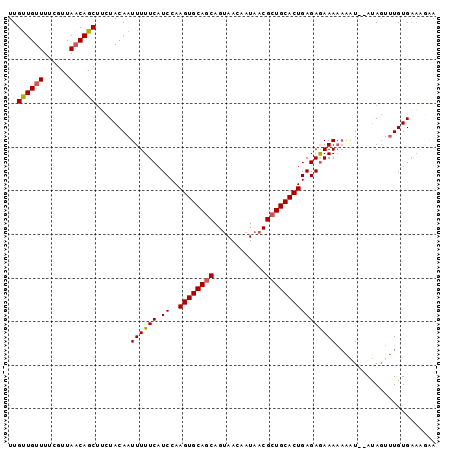
>3L_DroMel_CAF1 10768116 96 + 23771897 UUCUUUCACAAACUAU--AACAUUUUCUCUCAGUGCAGCGUUAUUGUUACUGCUGCACUUGGAUGAAAAAUUGUAGAAGCUGUUAACGAAAACAACAA ............((((--((..((((((((.(((((((((.((....)).))))))))).))).))))).))))))..(.((((......)))).).. ( -25.90) >DroSec_CAF1 101797 90 + 1 ------CACAAACUAU--AUUUUUUUUUCUCAGUGCAGCGUUAUUGUUACUGCUGCACUUGGAUGAAAAAUUGUAGAAGCUGUUAACGAAAACAACAA ------......((((--(.((((((.(((.(((((((((.((....)).))))))))).))).)))))).)))))..(.((((......)))).).. ( -27.20) >DroSim_CAF1 104690 96 + 1 UCCCUUCACAAACUAU--AUUUUUUUCUCUCAGUGCAGCGUUAUUGUUACUGCUGCACUUGGAUGAAAAAUUGUAGAAGCUGGUAACGAAAACAACAA ............((((--(.((((((.(((.(((((((((.((....)).))))))))).))).)))))).)))))...................... ( -26.10) >DroEre_CAF1 106703 95 + 1 UUGUUUCACAUAAUGUGUACAUGUUUCUCUCAGUGCACCGUUAUUGUUGCUGCUGCACUUGGAUGAAAAAUUGUA---GCUGUUAACGAAAACAGCAA ......(((.....)))((((..(((((((.(((((((.((.......)).).)))))).))).))))...))))---((((((......)))))).. ( -25.70) >consensus UUCUUUCACAAACUAU__AUUUUUUUCUCUCAGUGCAGCGUUAUUGUUACUGCUGCACUUGGAUGAAAAAUUGUAGAAGCUGUUAACGAAAACAACAA ............((((.....(((((((((.(((((((((.((....)).))))))))).))).))))))..))))..(.((((......)))).).. (-18.14 = -18.83 + 0.69)



| Location | 10,768,116 – 10,768,212 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -19.12 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975652 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10768116 96 - 23771897 UUGUUGUUUUCGUUAACAGCUUCUACAAUUUUUCAUCCAAGUGCAGCAGUAACAAUAACGCUGCACUGAGAGAAAAUGUU--AUAGUUUGUGAAAGAA ........(((....((((((...(((.((((((.((..((((((((.((.......)))))))))))))))))).))).--..)).))))....))) ( -24.80) >DroSec_CAF1 101797 90 - 1 UUGUUGUUUUCGUUAACAGCUUCUACAAUUUUUCAUCCAAGUGCAGCAGUAACAAUAACGCUGCACUGAGAAAAAAAAAU--AUAGUUUGUG------ ..((((((......))))))........((((((.((..((((((((.((.......)))))))))))))))))).....--..........------ ( -21.70) >DroSim_CAF1 104690 96 - 1 UUGUUGUUUUCGUUACCAGCUUCUACAAUUUUUCAUCCAAGUGCAGCAGUAACAAUAACGCUGCACUGAGAGAAAAAAAU--AUAGUUUGUGAAGGGA .......(..(.((((.((((.......((((((.((..((((((((.((.......))))))))))..))))))))...--..)))).)))).)..) ( -24.90) >DroEre_CAF1 106703 95 - 1 UUGCUGUUUUCGUUAACAGC---UACAAUUUUUCAUCCAAGUGCAGCAGCAACAAUAACGGUGCACUGAGAGAAACAUGUACACAUUAUGUGAAACAA ..((((((......))))))---((((...((((.((..((((((.(............).))))))..))))))..))))(((.....)))...... ( -23.10) >consensus UUGUUGUUUUCGUUAACAGCUUCUACAAUUUUUCAUCCAAGUGCAGCAGUAACAAUAACGCUGCACUGAGAGAAAAAAAU__AUAGUUUGUGAAAGAA ..((((((......))))))........((((((.((..((((((((............))))))))))))))))....................... (-19.12 = -19.25 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:41 2006