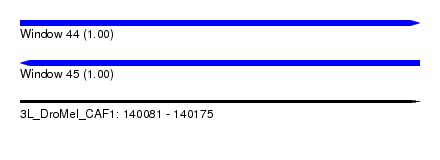
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 140,081 – 140,175 |
| Length | 94 |
| Max. P | 0.998595 |

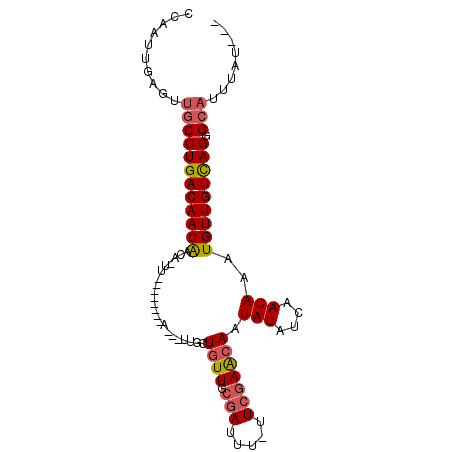
| Location | 140,081 – 140,175 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -13.34 |
| Energy contribution | -14.38 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 140081 94 + 23771897 ---AUAAAUGCCCUAACAACAUUUAUUGAUAUAUUCUUCGAAAAAAUCGCAACACCAACAAUCGCAACGCAACUAUCGUUGUCAGGCAACG-AAUGGG ---.....(((..........(((.((((........)))).)))...((.............))...))).(((((((((.....)))))-.)))). ( -12.82) >DroSec_CAF1 14430 78 + 1 ---AUAAAUGCCCUGACAACAUUUAUUGAUAUAUUGUUCGAA-AAAUCGCAACAGCAA----------------GUUGUUGUCAGGCAACUCAAUUGG ---.....((((.((((((((....(((.....((((((((.-...))).))))))))----------------..)))))))))))).......... ( -21.50) >DroSim_CAF1 14973 78 + 1 ---AUAAAUGCCCUGACAACAUUUAUUGAUAUAUUGUUCGAA-AAAUCGCAACAGCAA----------------GUUGUUGUCAGGCAACUCAAUUGG ---.....((((.((((((((....(((.....((((((((.-...))).))))))))----------------..)))))))))))).......... ( -21.50) >DroEre_CAF1 16028 80 + 1 UU--------CUCUGACAACAUUUAUGGAUAUAUUGCUUGAA-AAAUGGCAACAGCAAGUUU--------AAUUGUUGUUGUCAGGCAACAAAA-UGG ..--------.........(((((..................-...((((((((((((....--------..))))))))))))(....).)))-)). ( -19.70) >DroYak_CAF1 16469 89 + 1 UCCAUAAAUGCACUGACAACAUUUAUAGAUAUAUUGCUCGAAAAAAUCGCAACAGCAAGAUU--------AAUUGUUGUUGUCAGGCAACAAAA-UGC ........(((.(((((((((...((.(((...((((((((.....)))....))))).)))--------.))...))))))))))))......-... ( -23.50) >consensus ___AUAAAUGCCCUGACAACAUUUAUUGAUAUAUUGUUCGAA_AAAUCGCAACAGCAA___U________AA_UGUUGUUGUCAGGCAACAAAAUUGG ........(((.((((((((.....((((........)))).......((....)).....................))))))))))).......... (-13.34 = -14.38 + 1.04)

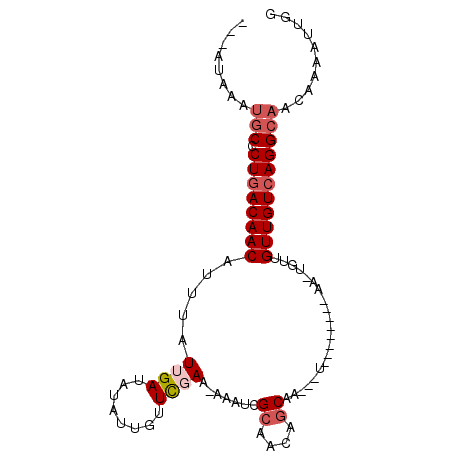
| Location | 140,081 – 140,175 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 76.10 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -15.20 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 140081 94 - 23771897 CCCAUU-CGUUGCCUGACAACGAUAGUUGCGUUGCGAUUGUUGGUGUUGCGAUUUUUUCGAAGAAUAUAUCAAUAAAUGUUGUUAGGGCAUUUAU--- ......-...((((((((((((((((((((...)))))))))(((((..(((.....)))......))))).......))))))).)))).....--- ( -24.90) >DroSec_CAF1 14430 78 - 1 CCAAUUGAGUUGCCUGACAACAAC----------------UUGCUGUUGCGAUUU-UUCGAACAAUAUAUCAAUAAAUGUUGUCAGGGCAUUUAU--- .....(((((.(((((((((((..----------------....((((.(((...-.))))))).(((....)))..)))))))).)))))))).--- ( -20.30) >DroSim_CAF1 14973 78 - 1 CCAAUUGAGUUGCCUGACAACAAC----------------UUGCUGUUGCGAUUU-UUCGAACAAUAUAUCAAUAAAUGUUGUCAGGGCAUUUAU--- .....(((((.(((((((((((..----------------....((((.(((...-.))))))).(((....)))..)))))))).)))))))).--- ( -20.30) >DroEre_CAF1 16028 80 - 1 CCA-UUUUGUUGCCUGACAACAACAAUU--------AAACUUGCUGUUGCCAUUU-UUCAAGCAAUAUAUCCAUAAAUGUUGUCAGAG--------AA ...-.........(((((((((.(((..--------....))).((((((.....-.....))))))..........)))))))))..--------.. ( -15.80) >DroYak_CAF1 16469 89 - 1 GCA-UUUUGUUGCCUGACAACAACAAUU--------AAUCUUGCUGUUGCGAUUUUUUCGAGCAAUAUAUCUAUAAAUGUUGUCAGUGCAUUUAUGGA ...-...((((((....(((((.(((..--------....))).)))))(((.....))).))))))..(((((((((((.......))))))))))) ( -22.20) >consensus CCAAUUGAGUUGCCUGACAACAACA_UU________A___UUGCUGUUGCGAUUU_UUCGAACAAUAUAUCAAUAAAUGUUGUCAGGGCAUUUAU___ ..........((((((((((((......................((((.(((.....))))))).(((....)))..))))))))).)))........ (-15.20 = -15.64 + 0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:27 2006