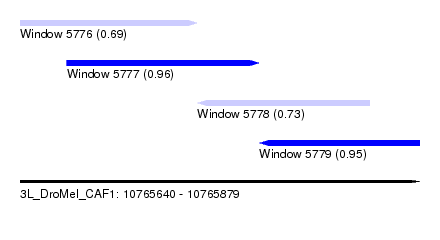
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,765,640 – 10,765,879 |
| Length | 239 |
| Max. P | 0.958421 |

| Location | 10,765,640 – 10,765,746 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -19.10 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694659 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10765640 106 + 23771897 AGACUCUUUCUCCCUGAUAGCUU------------UGUCUCCAUCGCACUCUUUCGCCAGAUCUCUGAAGCU--AGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGA .((((..(((((.(((((.((..------------((....))..)).((((..(..(((....)))..)..--))))))))).)))))..))))...(((....)))............ ( -29.20) >DroSec_CAF1 99330 108 + 1 AGACUCUUUCUCCCUGGUAGCUC------------CGUCUCCUUCGCACUCUUUCGCCAGAUCUCUGAAGCUAUAGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGA .((((..(((((.(((((.((..------------..........)).((((...(((((....)))..))...))))))))).)))))..))))...(((....)))............ ( -27.20) >DroSim_CAF1 102194 108 + 1 AGACUCUUUCUCCCUGGUAGCUC------------CGUCUCCUUCGCACUCUUUCGCCAGAUCUCCGAAGCUAUAGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGA .((((..(((((.(((((..(((------------..(...(((((...(((......)))....)))))..)..)))))))).)))))..))))...(((....)))............ ( -26.00) >DroEre_CAF1 104246 106 + 1 AGACUCUUUCUCCCAAGCAACUC------------CAUCUCCUUCGCACUCUUUCUCCAGAUCUCUGAAGCU--GGAGAUCAGUGAGAAUAAGUCAAAAGCAUGGGCAAUAAUAAAUAGA ...........((((.((.....------------...((..(((.((((...(((((((..........))--)))))..)))).)))..))......)).)))).............. ( -24.44) >DroYak_CAF1 115363 118 + 1 AGACUCUUUCUCCCAAGUAACUCCAUUUCCAUCUCCAUCUCCCUCGCACUCUUUCUCCAGAUCUCUGCAGCU--AGAGAUCGGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGA .((((..(((((((((((......))))...............................((((((((....)--))))))))).)))))..))))...(((....)))............ ( -25.00) >consensus AGACUCUUUCUCCCUGGUAGCUC____________CGUCUCCUUCGCACUCUUUCGCCAGAUCUCUGAAGCU__AGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGA .((((..(((((.(...............................((........))..(((((((........))))))).).)))))..))))...(((....)))............ (-19.10 = -19.78 + 0.68)


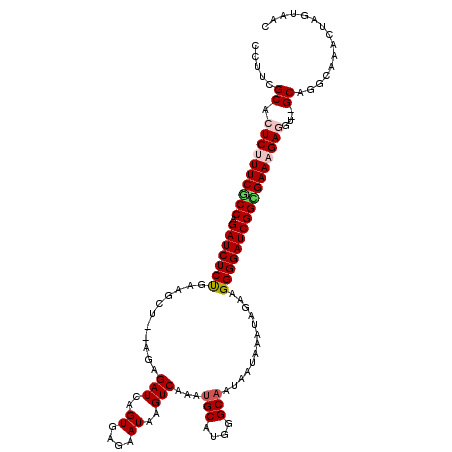
| Location | 10,765,668 – 10,765,783 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -34.88 |
| Consensus MFE | -26.76 |
| Energy contribution | -27.48 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10765668 115 + 23771897 CCAUCGCACUCUUUCGCCAGAUCUCUGAAGCU--AGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGAAGGGGAUCGGUGAACGAGGU--GCAGGCAAACUAGUAAC .....((((((.((((((.(((((((....((--(..(((..((....))..)))...(((....)))........)))..))))))))))))).)).))--))............... ( -30.40) >DroSec_CAF1 99358 119 + 1 CCUUCGCACUCUUUCGCCAGAUCUCUGAAGCUAUAGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGAAGGGGAUCGGCGAACGAGGUGUGCAGGCAAACUAGUAAC (((.(((((((.((((((.(((((((....((((...(((..((....))..)))...(((....))).......))))..))))))))))))).)).))))).)))............ ( -36.50) >DroSim_CAF1 102222 119 + 1 CCUUCGCACUCUUUCGCCAGAUCUCCGAAGCUAUAGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGAAGGGGAUCGGCGAAAGAGGUGUGCAGGCAAACUAGUAAC (((.((((((((((((((.(((((((....((((...(((..((....))..)))...(((....))).......))))..)))))))))))))))).))))).)))............ ( -42.50) >DroEre_CAF1 104274 115 + 1 CCUUCGCACUCUUUCUCCAGAUCUCUGAAGCU--GGAGAUCAGUGAGAAUAAGUCAAAAGCAUGGGCAAUAAUAAAUAGAAGGGGAUCGGUGAAAGAAGU--GCAGGCAAACUUGUAAC .((((.((((...(((((.(((((((......--)))))))..................((....))..............)))))..))))...))))(--(((((....)))))).. ( -30.90) >DroYak_CAF1 115403 115 + 1 CCCUCGCACUCUUUCUCCAGAUCUCUGCAGCU--AGAGAUCGGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGAAGGGGAUCGGAGAAAGAAGC--GCUGGAAAACUUGUAAC .((.(((..(((((((((.(((((((....((--(..(((..((....))..)))...(((....)))........)))..)))))))))))))))).))--)..))............ ( -34.10) >consensus CCUUCGCACUCUUUCGCCAGAUCUCUGAAGCU__AGAGAUCAGUGAGAAUAAGUCAAAUGCAUGGGCAAUAAUAAAUAGAAGGGGAUCGGCGAAAGAGGU__GCAGGCAAACUAGUAAC .....((.((((((((((.(((((((...........(((..((....))..)))...(((....))).............)))))))))))))))))....))............... (-26.76 = -27.48 + 0.72)


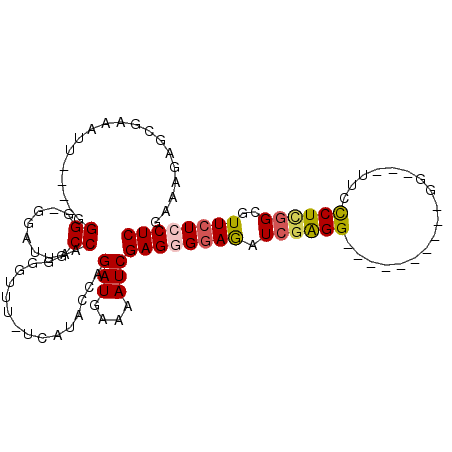
| Location | 10,765,746 – 10,765,849 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -35.44 |
| Consensus MFE | -23.71 |
| Energy contribution | -24.11 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10765746 103 - 23771897 AGGGGUUUCUCAUACCAGAUGAAAAUCGAGGGGAGAUCGAGG----------GGUUGUUCCCUCGGCGUUCUCCUCGUUACUAGUUUGCCUGC--ACCUCGUUCACCGAUCCCCU (((((..........(((((......(((((((((.((((((----------((....))))))))..)))))))))......))))).....--...(((.....))).))))) ( -38.20) >DroSec_CAF1 99438 102 - 1 AGGGGUUCUUCAUACCAGAUGAAAAUCGAGGGGAGAUCAAGG----------GG---UUCCCUCGGCGUUCUCCUCGUUACUAGUUUGCCUGCACACCUCGUUCGCCGAUCCCCU (((((.((.......(((((......(((((((((..(..((----------(.---...)))..)..)))))))))......)))))...((...........)).)).))))) ( -30.60) >DroSim_CAF1 102302 102 - 1 AGGGGUUUUUCAUAGAAGAUGGAAAUCGAGGGGAGAUCGAGG----------GG---UUCCCUCGGCGUUCUCCUCGUUACUAGUUUGCCUGCACACCUCUUUCGCCGAUCCCCU (((((....((...((((..((....(((((((((.((((((----------(.---..)))))))..)))))))))......((......))...))..))))...)).))))) ( -37.10) >DroEre_CAF1 104352 110 - 1 AGGGGUUU-UCAUACCAGAUGAAAAUCGAGGGAAAAUCGGGGGAAAAUCGAGGGG--UACCCUCGGAGUUCUGCUCGUUACAAGUUUGCCUGC--ACUUCUUUCACCGAUCCCCU (((((...-((......(((....)))(((((((...((((.(((...((((((.--..))))))(((.....)))........))).)))).--..)))))))...)).))))) ( -38.10) >DroYak_CAF1 115481 95 - 1 AGGGGUUU-UC-----AGAUGAAAAUCGAGGGGAGAUUGAGG----------GGU--UACUCUUCGAGUUCUGCUCGUUACAAGUUUUCCAGC--GCUUCUUUCUCCGAUCCCCU (((((...-((-----.(((....)))....(((((..((((----------.((--((((...((((.....)))).....))).....)))--.))))..))))))).))))) ( -33.20) >consensus AGGGGUUU_UCAUACCAGAUGAAAAUCGAGGGGAGAUCGAGG__________GG___UUCCCUCGGCGUUCUCCUCGUUACUAGUUUGCCUGC__ACCUCUUUCACCGAUCCCCU (((((....((......(((....)))((((((((.((((((..................))))))..))))))))...............................)).))))) (-23.71 = -24.11 + 0.40)



| Location | 10,765,783 – 10,765,879 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.95 |
| Mean single sequence MFE | -31.96 |
| Consensus MFE | -15.59 |
| Energy contribution | -15.99 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.49 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10765783 96 - 23771897 GAAAGAUCGAAAUUUGGUGGGG-GGAUUACCAGGGGUUUCUCAUACCAGAUGAAAAUCGAGGGGAGAUCGAGG----------GGUUGUUCCCUCGGCGUUCUCCUC ....(((....(((((((((((-..(((......)))..))).))))))))....)))((((((((.((((((----------((....))))))))..)))))))) ( -40.50) >DroSec_CAF1 99477 89 - 1 GAAAUAGCGAAAUG----GGGG-GGAUUACCAGGGGUUCUUCAUACCAGAUGAAAAUCGAGGGGAGAUCAAGG----------GG---UUCCCUCGGCGUUCUCCUC .............(----((((-((((..((..(((..((((...((.(((....)))...))(....)..))----------))---..)))..)).))))))))) ( -27.90) >DroSim_CAF1 102341 89 - 1 GAAAGAGCGAAAUU----GGGG-GGAUUACCAGGGGUUUUUCAUAGAAGAUGGAAAUCGAGGGGAGAUCGAGG----------GG---UUCCCUCGGCGUUCUCCUC ((((((.(....((----((..-......)))).).))))))......(((....)))((((((((.((((((----------(.---..)))))))..)))))))) ( -32.30) >DroEre_CAF1 104389 100 - 1 GAAAGAGCGAAAUU----GGGGGGGCUUACCAGGGGUUU-UCAUACCAGAUGAAAAUCGAGGGAAAAUCGGGGGAAAAUCGAGGGG--UACCCUCGGAGUUCUGCUC ....(((((((.((----.(((((...((((...(((((-((...((.(((....)))((.......))))..)))))))....))--))))))).)).))).)))) ( -33.20) >DroYak_CAF1 115518 84 - 1 GAAAGAGCGAAAUU----GGGG-GGAUUACCAGGGGUUU-UC-----AGAUGAAAAUCGAGGGGAGAUUGAGG----------GGU--UACUCUUCGAGUUCUGCUC ....(((((((.((----((((-((...(((...(((((-((-----.(((....)))....)))))))....----------)))--..)))))))).))).)))) ( -25.90) >consensus GAAAGAGCGAAAUU____GGGG_GGAUUACCAGGGGUUU_UCAUACCAGAUGAAAAUCGAGGGGAGAUCGAGG__________GG___UUCCCUCGGCGUUCUCCUC ...................((........)).................(((....)))((((((((.((((((..................))))))..)))))))) (-15.59 = -15.99 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:37 2006