| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,764,427 – 10,764,545 |
| Length | 118 |
| Max. P | 0.895282 |

| Location | 10,764,427 – 10,764,545 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -19.86 |
| Energy contribution | -20.14 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.895282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
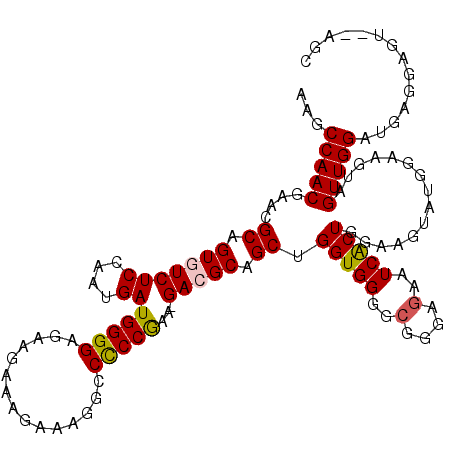
>3L_DroMel_CAF1 10764427 118 + 23771897 GCUCGACUCCUCAUCCAACUACUUCCAUACUUCCAGUGAUUCUCACGCCCCACCAGCUGCGUC-UUCGGAGGCUCUUCUUUCUUCUCCCCAUCACUGGAGACACUGCGUUCGUUGGCUU ..............(((((............(((((((((......((.......)).((.((-....)).)).................)))))))))(((.....))).)))))... ( -25.90) >DroSec_CAF1 98135 116 + 1 GCU--ACUCCUCAUCCAACUGCUUCCAUACUUCCAGUGAUUCUCCCCCACCACCAGCUGCGUC-UUCGGGGGCCUUUCUUUCUUCUCCCCAUCAUUGGAGACACUGCGUUCGUUGGCUU ...--...............(((..(..((...(((((.....(((((...(((....).)).-...)))))...........(((((........)))))))))).))..)..))).. ( -24.00) >DroSim_CAF1 101012 116 + 1 GCU--ACUCCUCAUCCAACUACUUCCAUACUUCCAGUGAUUCUCCCGCACCACCAGCUGCGUC-UUCGGGGGCCUUUCUUUCUUCUCCCCAUCAUUGGAGACACUGCGUUCGUUGGCUU ...--.........(((((............(((((((((.....((((.(....).))))..-...(((((.............))))))))))))))(((.....))).)))))... ( -26.82) >DroEre_CAF1 103075 108 + 1 GCCA--CUCUUCAUCCAACUAC--------UUUCAGCGAUUCUCCCGCCCCACCAGCUGCUUC-UUCGGGGGCCUUUCUUUCUUCUCCCCAUCAUUGGAGACACUGCGUUCGUUGGCUU ....--........(((((...--------.....(((.......)))...(((((..(((((-....)))))..........(((((........)))))..))).))..)))))... ( -23.50) >DroYak_CAF1 114135 117 + 1 GCCACACUCUUCAUCCAACUACUAC--UUCUUCCAGUGAUUCUCCCGCCCCACCAGCUGCUUCAUUUGGGGGCCUUUCUUUCUUCUCCCCAUCAUUGGAGACACUGCGUUCGUUGGCUU ..............(((((....((--......(((((........(((((..(((.((...)).))))))))..........(((((........)))))))))).))..)))))... ( -23.60) >consensus GCU__ACUCCUCAUCCAACUACUUCCAUACUUCCAGUGAUUCUCCCGCCCCACCAGCUGCGUC_UUCGGGGGCCUUUCUUUCUUCUCCCCAUCAUUGGAGACACUGCGUUCGUUGGCUU ..............(((((............(((((((((......((.......))..........(((((.............))))))))))))))(((.....))).)))))... (-19.86 = -20.14 + 0.28)


| Location | 10,764,427 – 10,764,545 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -23.34 |
| Energy contribution | -23.46 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10764427 118 - 23771897 AAGCCAACGAACGCAGUGUCUCCAGUGAUGGGGAGAAGAAAGAAGAGCCUCCGAA-GACGCAGCUGGUGGGGCGUGAGAAUCACUGGAAGUAUGGAAGUAGUUGGAUGAGGAGUCGAGC ...(((((..((...(((((((((....)))((((.............))))..)-)))))..(..((((..(....)..))))..)..........)).))))).............. ( -34.72) >DroSec_CAF1 98135 116 - 1 AAGCCAACGAACGCAGUGUCUCCAAUGAUGGGGAGAAGAAAGAAAGGCCCCCGAA-GACGCAGCUGGUGGUGGGGGAGAAUCACUGGAAGUAUGGAAGCAGUUGGAUGAGGAGU--AGC ...(((((.....(((((((((((....)))))).............((((((..-..(((.....))).)))))).....)))))......((....))))))).........--... ( -33.60) >DroSim_CAF1 101012 116 - 1 AAGCCAACGAACGCAGUGUCUCCAAUGAUGGGGAGAAGAAAGAAAGGCCCCCGAA-GACGCAGCUGGUGGUGCGGGAGAAUCACUGGAAGUAUGGAAGUAGUUGGAUGAGGAGU--AGC ...(((((.....((((((((((.....(((((...............)))))..-..((((.(....).)))))))))..)))))....(((....)))))))).........--... ( -33.06) >DroEre_CAF1 103075 108 - 1 AAGCCAACGAACGCAGUGUCUCCAAUGAUGGGGAGAAGAAAGAAAGGCCCCCGAA-GAAGCAGCUGGUGGGGCGGGAGAAUCGCUGAAA--------GUAGUUGGAUGAAGAG--UGGC ...(((((..((.(((((((((((....))))))............(((((...(-(......))...)))))........)))))...--------)).)))))........--.... ( -31.90) >DroYak_CAF1 114135 117 - 1 AAGCCAACGAACGCAGUGUCUCCAAUGAUGGGGAGAAGAAAGAAAGGCCCCCAAAUGAAGCAGCUGGUGGGGCGGGAGAAUCACUGGAAGAA--GUAGUAGUUGGAUGAAGAGUGUGGC ..((((....((......((((((((..(((((...............)))))..........(..((((..(....)..))))..).....--......)))))).))...)).)))) ( -30.06) >consensus AAGCCAACGAACGCAGUGUCUCCAAUGAUGGGGAGAAGAAAGAAAGGCCCCCGAA_GACGCAGCUGGUGGGGCGGGAGAAUCACUGGAAGUAUGGAAGUAGUUGGAUGAGGAGU__AGC ...(((((....((.(((((((....))(((((...............)))))...))))).)).(((((..(....)..)))))...............))))).............. (-23.34 = -23.46 + 0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:34 2006