| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,760,692 – 10,760,838 |
| Length | 146 |
| Max. P | 0.690983 |

| Location | 10,760,692 – 10,760,804 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -26.39 |
| Energy contribution | -27.64 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10760692 112 + 23771897 GGGGU-CGGCAACGCCAAAGUGUGAAGCGGUAACCGCUUUAAAGCGACUCGGGUCCACCCAAGUCGGCACCA-----CUACUCAGCUUAACCCAGCUGAACCGAGAUCGGGCCAAGAC-- ..((.-((....))))...((.((((((((...))))))))..))((((.(((....))).))))(((....-----....((((((......)))))).(((....)))))).....-- ( -43.70) >DroSec_CAF1 94420 110 + 1 AGGGU-CCGCAGCGCCAAAGUGUGAAGCGGUAACCGCUUUAAAGCGACUCGGGUCCACCCAAGACGGCUCCA-----CU---CAGCUUAACCCAGCUGAACCGAGAUCG-GCCAAGACAG .((..-(((.(((((.......((((((((...))))))))..))).)))))..))........((((((..-----.(---(((((......))))))...))).)))-.......... ( -38.30) >DroSim_CAF1 97249 101 + 1 AGGGU-CCGCAGCGCCAAAGUGUGAAGCGGUAACCGCUUUAAAGCGACUCAGGUCCACCCAAGUCGGCUCCA-----CU---CA----------GCUGAACCGAGAUCGGGCCAAGACUG ..(((-(((((((.....((((((((((((...)))))))).(((((((..((....))..)))).))).))-----))---..----------)))).........))))))....... ( -34.90) >DroEre_CAF1 99157 110 + 1 CGGACCCAACAAUGCAAAAGUGUGAAGCGGUAACCGCUUUAAAGCGACUCAGGUCCACCCAAGUCGGCUCCA-----CU---CAGCUUAACCCAGCUGAACCGAGAUCGGGCCAAGAG-- .((.(((...............((((((((...))))))))...(((((..((....))..))))).(((..-----.(---(((((......))))))...)))...))))).....-- ( -36.30) >DroYak_CAF1 110197 115 + 1 GGGGU-CCACAAUGCAAAAGUGUGAAGCGGUAACCGCUUUAAAGCGACUCAGGUCCACCCAAGUCGGCUCCACCACACU---CAGCUUAACCCAGGUGAACCGAGAUCGGGCCAAGACU- ..(((-((.....((...((((((((((((...))))))...(((((((..((....))..)))).)))....))))))---..))........((....))......)))))......- ( -37.00) >consensus AGGGU_CCGCAACGCCAAAGUGUGAAGCGGUAACCGCUUUAAAGCGACUCAGGUCCACCCAAGUCGGCUCCA_____CU___CAGCUUAACCCAGCUGAACCGAGAUCGGGCCAAGAC__ .((((..............((.((((((((...))))))))..))(((....))).)))).....(((..............(((((......)))))..(((....))))))....... (-26.39 = -27.64 + 1.25)

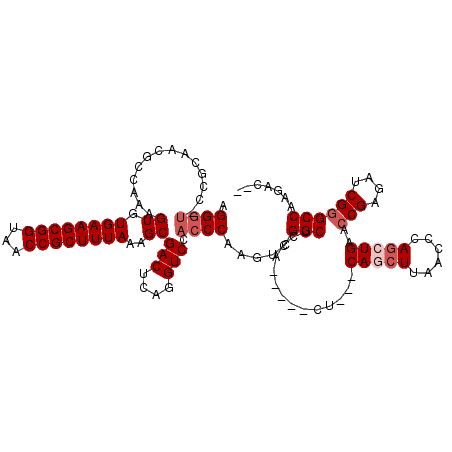

| Location | 10,760,692 – 10,760,804 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.73 |
| Mean single sequence MFE | -44.24 |
| Consensus MFE | -29.00 |
| Energy contribution | -30.16 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.510279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10760692 112 - 23771897 --GUCUUGGCCCGAUCUCGGUUCAGCUGGGUUAAGCUGAGUAG-----UGGUGCCGACUUGGGUGGACCCGAGUCGCUUUAAAGCGGUUACCGCUUCACACUUUGGCGUUGCCG-ACCCC --((((((..(((....)))..)))...(((...((..(((((-----(((((((((((((((....))))))))(((....))))).)))))))....)).)..))...))))-))... ( -46.60) >DroSec_CAF1 94420 110 - 1 CUGUCUUGGC-CGAUCUCGGUUCAGCUGGGUUAAGCUG---AG-----UGGAGCCGUCUUGGGUGGACCCGAGUCGCUUUAAAGCGGUUACCGCUUCACACUUUGGCGCUGCGG-ACCCU ..(.((..((-((....))))..))).(((((.(((.(---((-----((((((.(.((((((....)))))).)))))..((((((...))))))..)))))....)))...)-)))). ( -43.20) >DroSim_CAF1 97249 101 - 1 CAGUCUUGGCCCGAUCUCGGUUCAGC----------UG---AG-----UGGAGCCGACUUGGGUGGACCUGAGUCGCUUUAAAGCGGUUACCGCUUCACACUUUGGCGCUGCGG-ACCCU ..((((((..(((....)))..))((----------..---((-----((((((.((((..((....))..))))))))..((((((...))))))..))).)..)).....))-))... ( -39.40) >DroEre_CAF1 99157 110 - 1 --CUCUUGGCCCGAUCUCGGUUCAGCUGGGUUAAGCUG---AG-----UGGAGCCGACUUGGGUGGACCUGAGUCGCUUUAAAGCGGUUACCGCUUCACACUUUUGCAUUGUUGGGUCCG --.....(((((((((((((.....)))))....((.(---((-----((((((.((((..((....))..))))))))..((((((...))))))..)))))..))...)))))))).. ( -44.80) >DroYak_CAF1 110197 115 - 1 -AGUCUUGGCCCGAUCUCGGUUCACCUGGGUUAAGCUG---AGUGUGGUGGAGCCGACUUGGGUGGACCUGAGUCGCUUUAAAGCGGUUACCGCUUCACACUUUUGCAUUGUGG-ACCCC -.....((..(((....)))..))...(((((..((.(---((((((((((((((((((..((....))..))))(((....))))))).))))..)))))))..))......)-)))). ( -47.20) >consensus __GUCUUGGCCCGAUCUCGGUUCAGCUGGGUUAAGCUG___AG_____UGGAGCCGACUUGGGUGGACCUGAGUCGCUUUAAAGCGGUUACCGCUUCACACUUUGGCGUUGCGG_ACCCC ......((..(((....)))..))...((((...((............((((((.((((((((....))))))))))))))((((((...))))))..............))...)))). (-29.00 = -30.16 + 1.16)

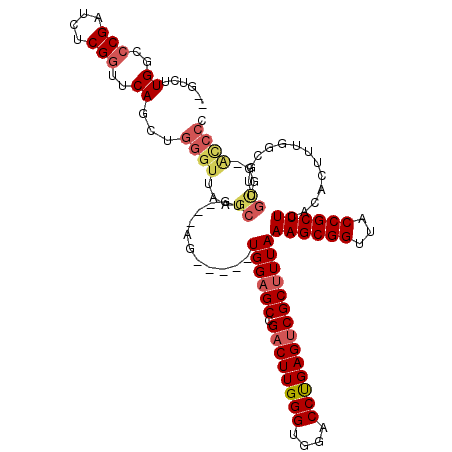

| Location | 10,760,731 – 10,760,838 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -35.56 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.11 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
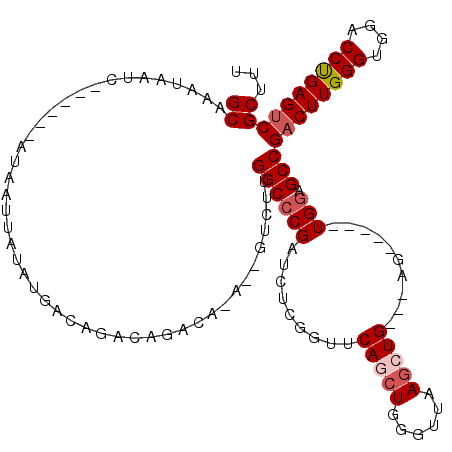
>3L_DroMel_CAF1 10760731 107 - 23771897 GCAAAUAAUC------AUAAUUAUAUGACAGACAGACAAA--GUCUUGGCCCGAUCUCGGUUCAGCUGGGUUAAGCUGAGUAG-----UGGUGCCGACUUGGGUGGACCCGAGUCGCUUU ((......((------(((....))))).((((.......--)))).((((((..(((((((.(((...))).)))))))...-----))).)))((((((((....))))))))))... ( -37.80) >DroSec_CAF1 94459 105 - 1 GCAAAUAAUC------AUAAUUAUAUGACAGACAGACAGACUGUCUUGGC-CGAUCUCGGUUCAGCUGGGUUAAGCUG---AG-----UGGAGCCGUCUUGGGUGGACCCGAGUCGCUUU ........((------(((....)))))(((((((.....))))).))((-((....))))((((((......)))))---).-----.(((((.(.((((((....)))))).)))))) ( -35.70) >DroSim_CAF1 97288 96 - 1 GCAAAUAAUC------AUAAUUAUAUGACAGACAGACAGACAGUCUUGGCCCGAUCUCGGUUCAGC----------UG---AG-----UGGAGCCGACUUGGGUGGACCUGAGUCGCUUU ((......((------(((....))))).....((((.....)))).((((((..(((((.....)----------))---))-----))).)))((((..((....))..))))))... ( -31.30) >DroEre_CAF1 99197 100 - 1 GCAAAUAAUC------AUAAUUAUAUGACAGCCAGU------CUCUUGGCCCGAUCUCGGUUCAGCUGGGUUAAGCUG---AG-----UGGAGCCGACUUGGGUGGACCUGAGUCGCUUU ........((------(((....)))))..(((((.------...)))))(((....)))(((((((......)))))---))-----.(((((.((((..((....))..))))))))) ( -34.50) >DroYak_CAF1 110236 112 - 1 GCAAAUAAUCAUAAUCAUAAUUAUAUGACAGGCGGC-----AGUCUUGGCCCGAUCUCGGUUCACCUGGGUUAAGCUG---AGUGUGGUGGAGCCGACUUGGGUGGACCUGAGUCGCUUU .........(((..(((((....)))))((.((..(-----((.(((((((((....)((....)).)))))))))))---.)).)))))((((.((((..((....))..)))))))). ( -38.50) >consensus GCAAAUAAUC______AUAAUUAUAUGACAGACAGACA_A__GUCUUGGCCCGAUCUCGGUUCAGCUGGGUUAAGCUG___AG_____UGGAGCCGACUUGGGUGGACCUGAGUCGCUUU ((.............................................((((((.........(((((......)))))..........))).)))((((((((....))))))))))... (-23.30 = -24.11 + 0.81)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:28 2006