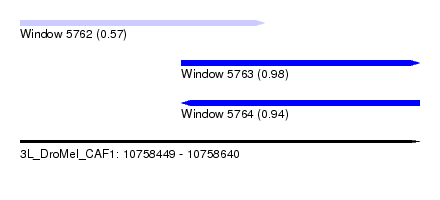
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,758,449 – 10,758,640 |
| Length | 191 |
| Max. P | 0.976068 |

| Location | 10,758,449 – 10,758,566 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 88.21 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -22.24 |
| Energy contribution | -25.04 |
| Covariance contribution | 2.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
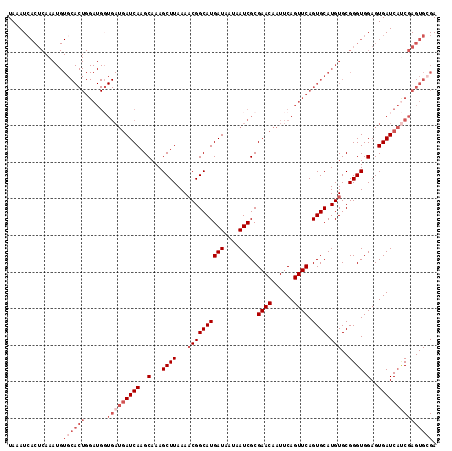
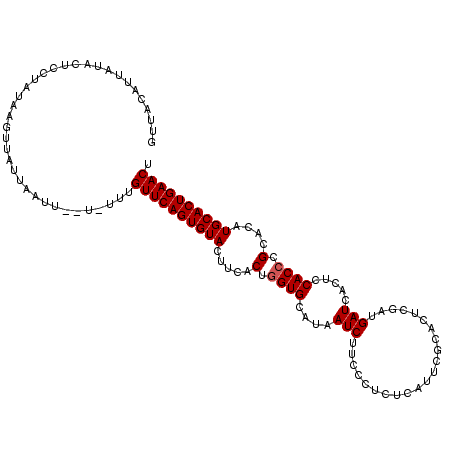
>3L_DroMel_CAF1 10758449 117 + 23771897 UAAAUAACUCAAAUGUGCACUGGAUGGUGAUGAUCAAGCAAAGCUUAAAACGGCAUGAUAAUAAUCGCGAACAAUUCAGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGA ......((((..(((((((((((((.(((((.((((.((...(.......).)).))))....)))))((.....)).)))))))))))))...(((((.....))))))))).... ( -32.40) >DroSec_CAF1 92182 117 + 1 UAAAUCACUCAAAUGUGCACUGGAUGGUGAUGAUCAAGCAAAGCUUAAAUCGGCAUGAUAAUAAUCGCGAACAAUUCAGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGA .....(((((..(((((((((((((.(((((.((((.((.............)).))))....)))))((.....)).)))))))))))))...(((((.....))))))))))... ( -34.42) >DroSim_CAF1 94995 117 + 1 UAAAUCACUCAAAUGUGCACUGGAUGGUGAUGAUCAAGCAAAGCUUAAAACGGCAUGAUAAUAAUCGCGAACAAUUCAGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGA .....(((((..(((((((((((((.(((((.((((.((...(.......).)).))))....)))))((.....)).)))))))))))))...(((((.....))))))))))... ( -34.50) >DroEre_CAF1 96899 91 + 1 UAAAUC--------------------------AUCAAGCAAAGCUUAAAACGGCAUGAUAGUAAUCGCGAACAAUUCAGUUCAGUGCAUGUGCGGGUGGAGUGAUCUGCGAGUGGGA ....((--------------------------((...(((..((((....(.(((((((....)))((((((......))))...))..)))).)...))))....)))..)))).. ( -18.60) >DroYak_CAF1 107881 117 + 1 UAAAUCACUCAAAUGUGCACUGGAUGCUGAUGAUCAAGCAAAGCUUAAAACGGCAUGAUAGUAAUCGCGAACAAUUCAGUUCAGUGCAUGUGCGGGUGGAGUGAUCACCGAGUGGGA ....((((((..(((((((((((..(((((.......((...(((......)))..(((....))))).......))))))))))))))))...(((((.....))))))))))).. ( -38.34) >consensus UAAAUCACUCAAAUGUGCACUGGAUGGUGAUGAUCAAGCAAAGCUUAAAACGGCAUGAUAAUAAUCGCGAACAAUUCAGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGA ...............((((((......(((((((((..(...((((...((((((((((....)))..((((......)))).)))).)))..)))).)..))))))))))))))). (-22.24 = -25.04 + 2.80)

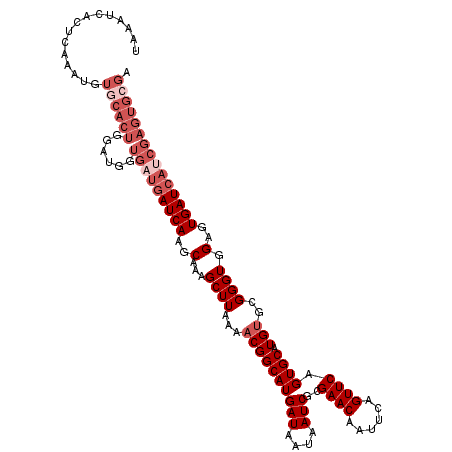
| Location | 10,758,526 – 10,758,640 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.976068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10758526 114 + 23771897 AGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGAAUGAGAGGUAAGAUUAUGCACCAGUGAAGUACACUGAACAAAAA--AAUUAAUAACUUACGGGAGUAUAUUGUAAC .((((((((.((..((.((((..((((((.(((....))).(....)....)))))).)))).))...)).)))))))).....--.....((((..(((....)))..))))... ( -29.60) >DroSec_CAF1 92259 113 + 1 AGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGAAUGAGAGGGAAGAUUAUGCACCAGUGAAGUACACUGAACAAA-A--AAUUAAUAUCUAAUAGAAGAAUAAUGUAAC .((((((((.((..((.((((..((((((.(((....))).(....)....)))))).)))).))...)).))))))))...-.--........(((......))).......... ( -25.80) >DroSim_CAF1 95072 113 + 1 AGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGAAUGAGAGGGAAGAUUAUGCACCAGUGAAGUACACUGAACAAA-A--AAUUAAUAAGUUAUAGGAGUAUAAUGUAAC .((((((((.((..((.((((..((((((.(((....))).(....)....)))))).)))).))...)).))))))))...-.--.........((((((.........)))))) ( -27.30) >DroEre_CAF1 96950 116 + 1 AGUUCAGUGCAUGUGCGGGUGGAGUGAUCUGCGAGUGGGAAUGAGAGGGAAGAUUAUGCACGAGUGAAGUACACUGAACAAAAAUAAAAAAAUAAAGAAGUGAAAUAUUAUCAAAA .((((((((.((..((..(((..(((((((.(..((....))..).....))))))).)))..))...)).))))))))..................................... ( -22.80) >consensus AGUUCAGUGCAUGUGCGGGUGGAGUGAUCAUCGAGUGCGAAUGAGAGGGAAGAUUAUGCACCAGUGAAGUACACUGAACAAA_A__AAUUAAUAACUAAUAGAAGUAUAAUGUAAC .((((((((.((..((.((((..((((((.((..((....))..)).....)))))).)))).))...)).))))))))..................................... (-22.48 = -22.98 + 0.50)


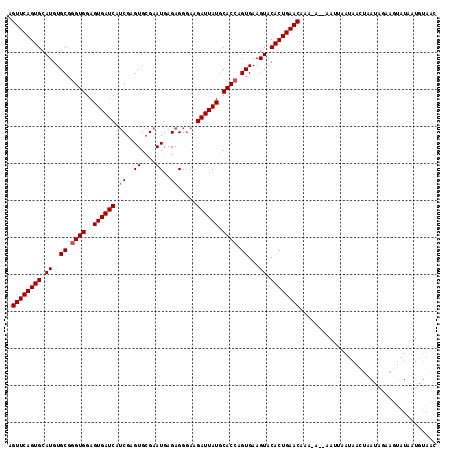
| Location | 10,758,526 – 10,758,640 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -17.70 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.78 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10758526 114 - 23771897 GUUACAAUAUACUCCCGUAAGUUAUUAAUU--UUUUUGUUCAGUGUACUUCACUGGUGCAUAAUCUUACCUCUCAUUCGCACUCGAUGAUCACUCCACCCGCACAUGCACUGAACU .....(((.(((....))).))).......--.....(((((((((((......)((((.............((((.((....))))))...........)))).)))))))))). ( -18.95) >DroSec_CAF1 92259 113 - 1 GUUACAUUAUUCUUCUAUUAGAUAUUAAUU--U-UUUGUUCAGUGUACUUCACUGGUGCAUAAUCUUCCCUCUCAUUCGCACUCGAUGAUCACUCCACCCGCACAUGCACUGAACU .....((((.(((......)))...)))).--.-...(((((((((((......)((((.............((((.((....))))))...........)))).)))))))))). ( -18.95) >DroSim_CAF1 95072 113 - 1 GUUACAUUAUACUCCUAUAACUUAUUAAUU--U-UUUGUUCAGUGUACUUCACUGGUGCAUAAUCUUCCCUCUCAUUCGCACUCGAUGAUCACUCCACCCGCACAUGCACUGAACU ..............................--.-...(((((((((((......)((((.............((((.((....))))))...........)))).)))))))))). ( -18.55) >DroEre_CAF1 96950 116 - 1 UUUUGAUAAUAUUUCACUUCUUUAUUUUUUUAUUUUUGUUCAGUGUACUUCACUCGUGCAUAAUCUUCCCUCUCAUUCCCACUCGCAGAUCACUCCACCCGCACAUGCACUGAACU .....................................((((((((((........((((...........((((..........).)))...........)))).)))))))))). ( -14.35) >consensus GUUACAUUAUACUCCUAUAAGUUAUUAAUU__U_UUUGUUCAGUGUACUUCACUGGUGCAUAAUCUUCCCUCUCAUUCGCACUCGAUGAUCACUCCACCCGCACAUGCACUGAACU .....................................((((((((((.....(.((((....(((......................))).....)))).)....)))))))))). (-15.52 = -15.78 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:23 2006