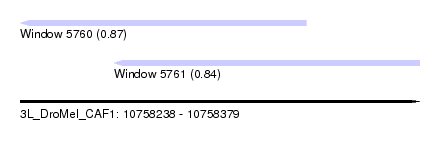
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,758,238 – 10,758,379 |
| Length | 141 |
| Max. P | 0.872466 |

| Location | 10,758,238 – 10,758,339 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.32 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -24.00 |
| Energy contribution | -25.44 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10758238 101 - 23771897 AUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACU------------GACUGCACUGAGUUGCAUAAUUUAAUAAGCGAGGUUGGAUUCAAUAGCUUCUAUUAACCUAC ......((((((((((((((.....((.....)).....------------).)))))).))))))).....(((((((((...((((....)))).)))..))))))..... ( -23.80) >DroSec_CAF1 91983 89 - 1 AUUUUCGCAAUUUGUGCAGCUU--------------------UG----GGGGGCUGCACUGAGUUGCAUAAAUUAAUAAGCGAGGUUGGAUUCAAUAGCUUCUAUUAACUGAC ......((((((((((((((((--------------------..----..))))))))).))))))).....(((((((((...((((....)))).)))..))))))..... ( -28.50) >DroSim_CAF1 94773 112 - 1 AUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUGGUUGAAU-GGGGGCUGCACUGAGUUGCAUAAAUUAAUAAGCGAGGUUGGAUUCAAUAGCUUCUAUUAACUGAC .((((.(((((((((((((((.((......((((.....))))....-)).)))))))).))))))).))))(((((((((...((((....)))).)))..))))))..... ( -35.60) >DroEre_CAF1 96677 113 - 1 AUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUGCUUGAAUGGGGGCCUGCACUGAGUUGCAUAAAUUAAUAAGCGAGGUUCGAUUCAAUAGCUUCUAUUAGAUGAU .((((.(((((((((((((((.((.(((.((......)).)))....)).)).)))))).))))))).))))((((((...((((((.........))))))))))))..... ( -28.00) >DroYak_CAF1 107658 113 - 1 AUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUGCUUGAAUGGGGGGCUGCACUGAGUUGCAUAAAUUAAUAAGCGAGGUUCGAUUCAAUAGCUUCUAUUAAGAGGC .((((((((((((((((((((.((((((.((......)).)))....))).)))))))).))))))).((......)).))))).............((((((....)))))) ( -35.70) >consensus AUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUG_UUGAAU_GGGGGCUGCACUGAGUUGCAUAAAUUAAUAAGCGAGGUUGGAUUCAAUAGCUUCUAUUAACUGAC ......(((((((((((((((.((........................)).)))))))).))))))).....((((((...(((((((.......)))))))))))))..... (-24.00 = -25.44 + 1.44)

| Location | 10,758,271 – 10,758,379 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.32 |
| Mean single sequence MFE | -28.07 |
| Consensus MFE | -21.00 |
| Energy contribution | -22.40 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835312 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10758271 108 - 23771897 UUGCGCAAUUUUAAUAAAUAUUUACAUUUGUUUCAAACCAAUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACU------------GACUGCACUGAGUUGCAUAAUUUAAUAAG ((((((((....(((((((......)))))))..........)))))))).((((((((((...((...((.((.....------------)))).))...))))))))))......... ( -23.74) >DroSec_CAF1 92016 96 - 1 UUGCGCAAUUUUAAUAAAUAUUUACAUUUGUUUCAAACCAAUUUUCGCAAUUUGUGCAGCUU--------------------UG----GGGGGCUGCACUGAGUUGCAUAAAUUAAUAAG ............(((((((......))))))).......(((((..((((((((((((((((--------------------..----..))))))))).)))))))..)))))...... ( -23.90) >DroSim_CAF1 94806 119 - 1 UUGCGCAAUUUUAAUAAAUAUUUACAUUUGUUUCAAACCAAUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUGGUUGAAU-GGGGGCUGCACUGAGUUGCAUAAAUUAAUAAG ....((((....(((((((......)))))))..........))))(((((((((((((((.((......((((.....))))....-)).)))))))).)))))))............. ( -32.84) >DroEre_CAF1 96710 120 - 1 UUGCGCAAUUUUAAUAAAUAUUUACAUUUGUUUCAAACCAAUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUGCUUGAAUGGGGGCCUGCACUGAGUUGCAUAAAUUAAUAAG ....((((....(((((((......)))))))..........))))(((((((((((((((.((.(((.((......)).)))....)).)).)))))).)))))))............. ( -27.64) >DroYak_CAF1 107691 120 - 1 UUGCGCAAUUUUAAUAAAUAUUUACAUUUGUUUCAAACCAAUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUGCUUGAAUGGGGGGCUGCACUGAGUUGCAUAAAUUAAUAAG ....((((....(((((((......)))))))..........))))(((((((((((((((.((((((.((......)).)))....))).)))))))).)))))))............. ( -32.24) >consensus UUGCGCAAUUUUAAUAAAUAUUUACAUUUGUUUCAAACCAAUUUGCGCAAUUUGUGCAGCUGCCUGAGAAGAUCUAACUG_UUGAAU_GGGGGCUGCACUGAGUUGCAUAAAUUAAUAAG ....((((....(((((((......)))))))..........))))(((((((((((((((.((........................)).)))))))).)))))))............. (-21.00 = -22.40 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:20 2006