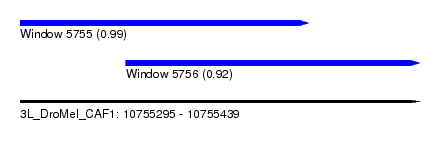
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,755,295 – 10,755,439 |
| Length | 144 |
| Max. P | 0.994746 |

| Location | 10,755,295 – 10,755,399 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.59 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -25.80 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10755295 104 + 23771897 UUCCCAGCUUUUAUCCCAUCA--UAGCCUGGGAACCUUUUCGCCU---------UUAGCUUCAGCUCUCGAAUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGAG-----UUCCAUAAAU (((((((((............--.)).)))))))....(((((..---------..(((....((((.((((((((....)).)))))).))))...))))))))-----.......... ( -33.12) >DroSec_CAF1 89058 104 + 1 UUCCCACCUUUUAUCCCAUCA--UGGCCUGGGAACCUUUUCGCCU---------UUAGCUUCAGCUCUCGAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGGA-----GUCCAUAAAU ((((((((.............--.))..))))))...((((((..---------..(((....((((.((((((((....)).)))))).))))...))))))))-----)......... ( -30.84) >DroSim_CAF1 91591 104 + 1 UUCCCACCUUUUAUCCCAUCA--UAGCCUGGGAACCUUUUCGCCU---------UUAGCUUCAGCUCUCGAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGGA-----GUCCAUAAAU .............(((((...--.....)))))....((((((..---------..(((....((((.((((((((....)).)))))).))))...))))))))-----)......... ( -30.10) >DroEre_CAF1 93744 104 + 1 UUCCCACCUUUUAUGCCAUCA--UAGCCUGGGAACCUUUUCGCCU---------UUAGCUGUAUCUCUCCAGUGCCUCAAGGUCAUUGGCGAGCGUUGCUGCGAG-----GUCUAUAAAU ((((((....(((((....))--)))..)))))).......((((---------(((((.....(((.((((((((....)).)))))).)))....)))).)))-----))........ ( -30.70) >DroYak_CAF1 104579 120 + 1 UUCCCACCUUUUAUCCCAUCAUAUAGCCUGGGAACCUUUUCGCCUUUUCGCCUUUUAGCUUUAGCUCUCCAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGAGCGUUGGUCUAUAAAU ((((((..((.(((......))).))..)))))).......(((....(((.((.((((....((((.(.((((((....)).)))).).))))...)))).)))))..)))........ ( -29.30) >consensus UUCCCACCUUUUAUCCCAUCA__UAGCCUGGGAACCUUUUCGCCU_________UUAGCUUCAGCUCUCGAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGAG_____GUCCAUAAAU ((((((......................))))))....(((((.............(((....((((.((((((((....)).)))))).))))...))))))))............... (-25.80 = -25.32 + -0.48)

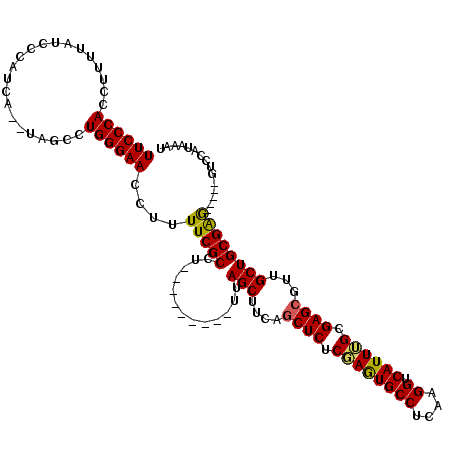
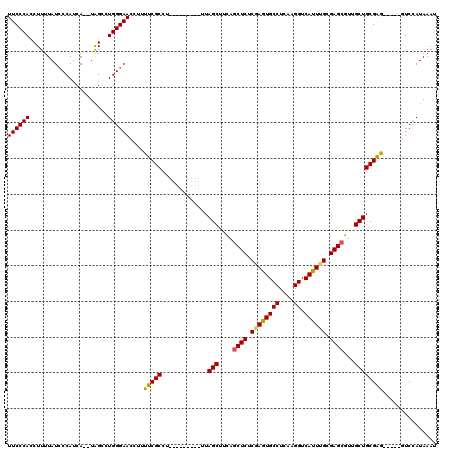
| Location | 10,755,333 – 10,755,439 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.07 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -25.69 |
| Energy contribution | -25.69 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10755333 106 + 23771897 CGCCU---------UUAGCUUCAGCUCUCGAAUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGAG-----UUCCAUAAAUAUGCAUGUCUGAUAUGUCGGCAUAUUACAAUACUUAUCGC .....---------..(((....((((.((((((((....)).)))))).))))...)))((((.-----.......(((((((.((.(......).)))))))))..........)))) ( -29.37) >DroSec_CAF1 89096 106 + 1 CGCCU---------UUAGCUUCAGCUCUCGAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGGA-----GUCCAUAAAUAUGCAUGUCUGAUAUGUCGGCAUAUUACAAUACUUAUCGC (((..---------..(((....((((.((((((((....)).)))))).))))...))))))((-----((.....(((((((.((.(......).))))))))).....))))..... ( -30.50) >DroSim_CAF1 91629 106 + 1 CGCCU---------UUAGCUUCAGCUCUCGAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGGA-----GUCCAUAAAUAUGCAUGUCUAAUAUGUCGGCAUAUUACAAUACUUAUCGC (((..---------..(((....((((.((((((((....)).)))))).))))...))))))((-----((.....(((((((.((.(......).))))))))).....))))..... ( -30.50) >DroEre_CAF1 93782 106 + 1 CGCCU---------UUAGCUGUAUCUCUCCAGUGCCUCAAGGUCAUUGGCGAGCGUUGCUGCGAG-----GUCUAUAAAUAUGCAUGUCUGAUAUGGCGGCAUAUUACAAUACUUAUCUC .((((---------(((((.....(((.((((((((....)).)))))).)))....)))).)))-----)).....(((((((.((((......))))))))))).............. ( -33.80) >DroYak_CAF1 104619 120 + 1 CGCCUUUUCGCCUUUUAGCUUUAGCUCUCCAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGAGCGUUGGUCUAUAAAUAUGCAUGUCUGAUAUGGCGGCAUAUUACAAUACUUAUCUC .(((....(((.((.((((....((((.(.((((((....)).)))).).))))...)))).)))))..))).....(((((((.((((......))))))))))).............. ( -34.00) >consensus CGCCU_________UUAGCUUCAGCUCUCGAGUGCCUCAAGGUCAUUUGCGAGCGUUGCUGCGAG_____GUCCAUAAAUAUGCAUGUCUGAUAUGUCGGCAUAUUACAAUACUUAUCGC (((.............(((....((((.((((((((....)).)))))).))))...))))))..............(((((((.((.(......).))))))))).............. (-25.69 = -25.69 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:15 2006