
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,750,945 – 10,751,086 |
| Length | 141 |
| Max. P | 0.728094 |

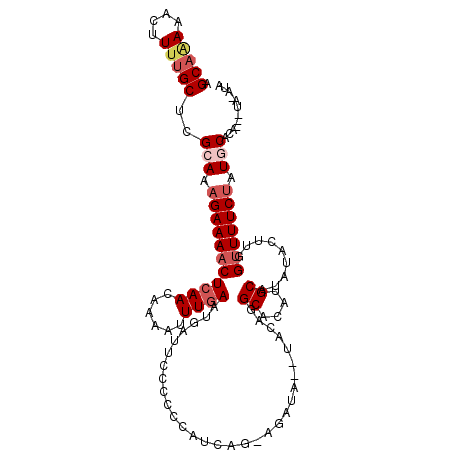
| Location | 10,750,945 – 10,751,047 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 84.32 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -11.58 |
| Energy contribution | -12.86 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10750945 102 - 23771897 AGCAAAAACUUUUGCUCGCAAAGAAAACUCAACAAAAUUUGAAUGAUUCCCCCCUUCAUCAGAUA--UACAGGCACAUGCAUAUACUUGGUUUUCUAUGCACA---U----- ((((((....)))))).(((.(((((((.(((....((((((.(((.........))))))))).--.....((....))......)))))))))).)))...---.----- ( -22.70) >DroSec_CAF1 84330 105 - 1 AGCAAAAACUUUUGCUCGCAAAGAAAACUCAACAAAAUUUGAAUGAUUCCCCCCACCAG-AGAU---UACAGGCGCAUGCAUAUACUUGGUUUUCUAUGCACA---UACAUA ((((((....)))))).(((.(((((((.(((...........((((..(........)-..))---))...((....))......)))))))))).)))...---...... ( -17.60) >DroSim_CAF1 87023 106 - 1 AGCAAAAACUUUUGCUCGCAAAGAAAACUCAACAAAAUUUGAAUGAUUCCCCCCAUCAG-AGAUA--UUCAGGCGCAAGCAUAUACUUGGUUUUCUAUGCACA---UACAUA ((((((....)))))).(((.(((((((.(((......(((((((.(((.........)-)).))--)))))((....))......)))))))))).)))...---...... ( -26.10) >DroEre_CAF1 89279 97 - 1 AGCAAAAACUUGUGCGCGCAAAGAAAACUCAACAA-AUUUGAAUGAUUCC--CCAUCAG-AUAU----ACAGGCACAUGCAUAUAGUUAGUUUUCUUUACACA-------UA .(((........)))(.(.((((((((((.(((..-((((((.((.....--.))))))-))..----....((....)).....))))))))))))).).).-------.. ( -19.40) >DroYak_CAF1 99988 109 - 1 AGCUGAAACUUUUGCUCGCAAAGAAAACUCAACAA-AUUUCAAUGAUUCCC-CCAUCAG-AUAUAUGUACAAGCACAUGCAUAUAGUUCGGUUUCCAUACACAACCUAUAUA ....((((((((((....))))(((..........-.......((((....-..)))).-.((((((((........)))))))).)))))))))................. ( -13.30) >consensus AGCAAAAACUUUUGCUCGCAAAGAAAACUCAACAAAAUUUGAAUGAUUCCCCCCAUCAG_AGAUA__UACAGGCACAUGCAUAUACUUGGUUUUCUAUGCACA___UA_AUA .(((((....)))))..(((.(((((((((((......))))..............................((....)).........))))))).)))............ (-11.58 = -12.86 + 1.28)

| Location | 10,750,977 – 10,751,086 |
|---|---|
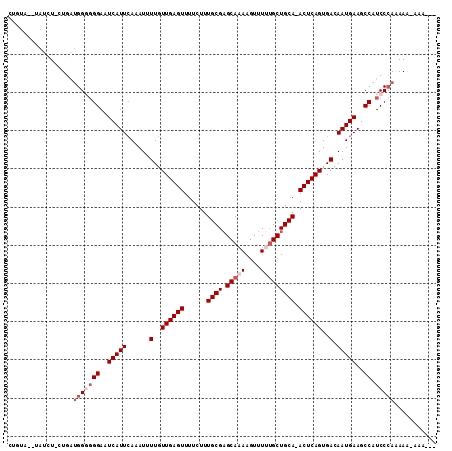
| Length | 109 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -24.90 |
| Consensus MFE | -19.98 |
| Energy contribution | -21.58 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.604099 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10750977 109 + 23771897 CUGUA--UAUCUGAUGAAGGGGGGAAUCAUUCAAAUUUUGUUGAGUUUUCUUUGCGAGCAAAAGUUUUUGCUGCA-ACUCAGUGACAAUGAAGCCAUCCAAAAAAAAAAGAA .....--...........((..((..(((((......(..((((((......(((.((((((....)))))))))-))))))..).)))))..))..))............. ( -23.20) >DroSec_CAF1 84367 104 + 1 CUGUA---AUCU-CUGGUGGGGGGAAUCAUUCAAAUUUUGUUGAGUUUUCUUUGCGAGCAAAAGUUUUUGCUGCA-ACUCAGUGACAAUGAAGCCAUCCCAAAAAAAAA--- .....---....-....(((((((..(((((......(..((((((......(((.((((((....)))))))))-))))))..).)))))..)).)))))........--- ( -26.40) >DroSim_CAF1 87060 105 + 1 CUGAA--UAUCU-CUGAUGGGGGGAAUCAUUCAAAUUUUGUUGAGUUUUCUUUGCGAGCAAAAGUUUUUGCUGCA-ACUCAGUGACAAUGAAGCCAUCCCAAAAACAAA--- .....--.....-....(((((((..(((((......(..((((((......(((.((((((....)))))))))-))))))..).)))))..)).)))))........--- ( -26.50) >DroEre_CAF1 89312 95 + 1 CUGU----AUAU-CUGAUGG--GGAAUCAUUCAAAU-UUGUUGAGUUUUCUUUGCGCGCACAAGUUUUUGCUGCA-ACUCAGUGACAAUGAAGCCAUCCCACAA-------- ....----....-....(((--(((....((((...-(..((((((......((((.(((........)))))))-))))))..)...))))....)))).)).-------- ( -22.40) >DroYak_CAF1 100028 101 + 1 UUGUACAUAUAU-CUGAUGG-GGGAAUCAUUGAAAU-UUGUUGAGUUUUCUUUGCGAGCAAAAGUUUCAGCUGCAAACUCAGUGACAAUGAAGCCAUCCCACAA-------- ............-.((.(((-(((..((((((....-(..(((((((.....(((.(((..........)))))))))))))..)))))))..))..)))))).-------- ( -26.00) >consensus CUGUA__UAUCU_CUGAUGGGGGGAAUCAUUCAAAUUUUGUUGAGUUUUCUUUGCGAGCAAAAGUUUUUGCUGCA_ACUCAGUGACAAUGAAGCCAUCCCAAAAA_AAA___ .................(((((((..(((((......(..((((((......((((.(((((....))))))))).))))))..).)))))..)).)))))........... (-19.98 = -21.58 + 1.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:12 2006