
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,749,106 – 10,749,248 |
| Length | 142 |
| Max. P | 0.972677 |

| Location | 10,749,106 – 10,749,225 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 90.57 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -29.88 |
| Energy contribution | -29.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972677 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10749106 119 + 23771897 AACAGCGAGUCAUUCGCUUGUGUCCGAUCAUCGAAAGCCGCAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGCGGCCCACAAAGAAUGCAUUUGCAUGUGUCCGGCAUCAACA .((((((((((((((..(((((..((.....))...(((((.((...((((((((..(....)..))))))))..))))))).))))).))))).)))))).))).............. ( -40.30) >DroSec_CAF1 82578 118 + 1 AACAGCGAGUCAUUCGCUUGUGUCCGAUCAUCGAAAGCUGCAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGUGGCCCACA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACA .((((((((((((((...((((..((.....))...((..(.((...((((((((..(....)..))))))))..)))..)).))))-.))))).)))))).))).............. ( -33.80) >DroSim_CAF1 85222 111 + 1 AA-------UCAUUCGCUUGUGUCCGAUCAUCGAAAGCCGCAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGUGGCCCACA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACA ..-------......(.(((((.(((..........(((((.((...((((((((..(....)..))))))))..)))))))..(((-...((((....)))).))).))))).)))). ( -30.70) >DroEre_CAF1 87494 111 + 1 -------UCCGAACAGCUAGUGUCCGAUCAUCGAAAGCCGCAUCAUUAUGACCAUUGUCAUGGUGGUGAUCAUUAGAGUGGCCCAGA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACA -------.(((.(((((.(((((....((.((....((((((((((((((((....)))))))))))..((....)))))))...))-.))..))))).)).)))...)))........ ( -31.30) >DroYak_CAF1 98168 111 + 1 -------AGUCAUUCGGUUGUGUCCGAUCAUCGAAAGCCGCAUCAUUAUGAUCAUUGUCAUGGUGGUGAUCAUUAGAGUGGCCCACA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACA -------.........((((((.(((..........(((((.((...((((((((..(....)..))))))))..)))))))..(((-...((((....)))).))).))))).)))). ( -35.80) >consensus AA_____AGUCAUUCGCUUGUGUCCGAUCAUCGAAAGCCGCAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGUGGCCCACA_AGAAUGCAUUUGCAUGUGUCCGGCAUCAACA ...................(((.(((..(((.....(((((.((...((((((((..(....)..))))))))..))))))).........((((....)))))))..))))))..... (-29.88 = -29.76 + -0.12)

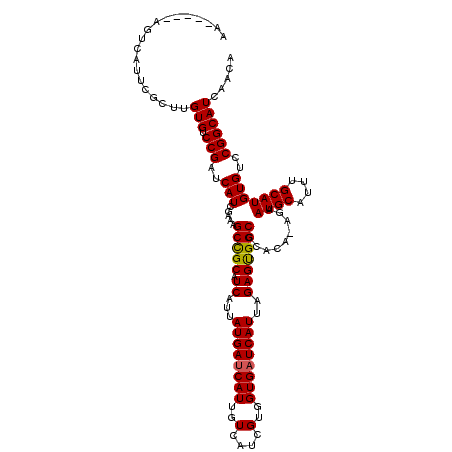
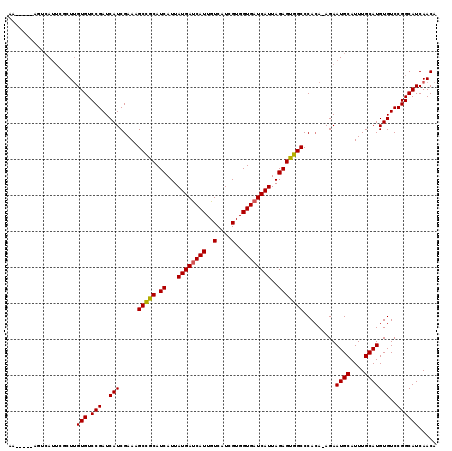
| Location | 10,749,146 – 10,749,248 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 88.17 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -21.26 |
| Energy contribution | -21.14 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10749146 102 + 23771897 CAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGCGGCCCACAAAGAAUGCAUUUGCAUGUGUCCGGCAUCAACAGCUCCAUUUCCAUUU------CCAUUUCC----------- .......((((((((..(....)..))))))))..((((.(((.(((....((((....)))).)))..))).......))))...........------........----------- ( -23.20) >DroSec_CAF1 82618 95 + 1 CAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGUGGCCCACA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACAGCUCCAUUUCCAU------------UUCC----------- .......((((((((..(....)..))))))))..((((.(((.(((-...((((....)))).)))..))).......)))).........------------....----------- ( -22.20) >DroSim_CAF1 85255 107 + 1 CAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGUGGCCCACA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACAGCUCCAUUUCCAUUUCCAAUUCCAUUUCC----------- .......((((((((..(....)..))))))))..((((.(((.(((-...((((....)))).)))..))).......)))).........................----------- ( -22.20) >DroEre_CAF1 87527 101 + 1 CAUCAUUAUGACCAUUGUCAUGGUGGUGAUCAUUAGAGUGGCCCAGA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACAGCUCCAUUACCAUUU------CCAUGUUC----------- ((((((((((((....)))))))))))).......((((.(((..((-...((((....))))...)).))).......))))...........------........----------- ( -26.80) >DroYak_CAF1 98201 112 + 1 CAUCAUUAUGAUCAUUGUCAUGGUGGUGAUCAUUAGAGUGGCCCACA-AGAAUGCAUUUGCAUGUGUCCGGCAUCAACAGCUCCAUUUCCAUUU------CCAUUUUCAUUUCCAUUUC ((((((((((((....)))))))))))).......((((.(((.(((-...((((....)))).)))..))).......))))...........------................... ( -25.30) >consensus CAUCAUUAUGAUCAUUGUCAUCGUGGUGAUCAUUAGAGUGGCCCACA_AGAAUGCAUUUGCAUGUGUCCGGCAUCAACAGCUCCAUUUCCAUUU______CCAUUUCC___________ (((((..(((((....)))))..))))).......((((.(((.(((....((((....)))).)))..))).......)))).................................... (-21.26 = -21.14 + -0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:10 2006