
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,748,229 – 10,748,411 |
| Length | 182 |
| Max. P | 0.998353 |

| Location | 10,748,229 – 10,748,331 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.19 |
| Mean single sequence MFE | -33.81 |
| Consensus MFE | -24.41 |
| Energy contribution | -24.63 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10748229 102 + 23771897 GCUGCUCAA-AGUGCAGUGCGGCUCAAAACAAAGUUAUUACGGCCUAAG------------CUUUGUUUGCCUUUCUGGCUCAUGGCAAA-----AAACCCACAUGGCCAGAUCGGCCCC (((((.(..-......).)))))...((((((((((...........))------------))))))))(((..((((((.((((.....-----.......))))))))))..)))... ( -35.50) >DroSec_CAF1 81729 102 + 1 GCUGCUCAA-ACUGCAGUGCGGCUCAAAACAAAGUUAUUACGGCCUAAG------------CUUUGUUUGCCUUUCUGGCUCAUGGCAAA-----AAACCCACAUGACCAGAUCGGCCCC (((((.(..-......).)))))...((((((((((...........))------------))))))))(((..(((((.(((((.....-----.......))))))))))..)))... ( -33.40) >DroSim_CAF1 84355 102 + 1 GCUGCUCAA-ACUGCAGUGCGGCUCAAAACAAAGUUAUUACGGCCUAAG------------CUUUGUUUGCCUUUCUGGCUCAUGGCAAA-----AAACCCACAUGGCCAGAUCGGCCCC (((((.(..-......).)))))...((((((((((...........))------------))))))))(((..((((((.((((.....-----.......))))))))))..)))... ( -35.50) >DroEre_CAF1 86635 104 + 1 GCAGCACAAAACUGCAGUGCGGCCCAAAACAAAGUUAUUACGGCCUAAG------------CUUUGUUUGCCUUUCUGGCUCAUGGCAAAA----AAACCCACAUGGCCAGAUCGGCCCC ((.((((.........)))).))...((((((((((...........))------------))))))))(((..((((((.((((......----.......))))))))))..)))... ( -34.32) >DroYak_CAF1 97278 107 + 1 GCUGCUCAA-AGUGCAGUGCGGCCCAAAACAAAGUUAUUACGGCCUAAG------------CUUUGUUUGCCUUUCUGGCUCAUGGCAAAAAAAAAAACCCACAUGGCCAGAUCGGCCCC (((((....-...)))))..((((..((((((((((...........))------------)))))))).....((((((.((((.................))))))))))..)))).. ( -35.23) >DroAna_CAF1 84136 112 + 1 GC--CUCAA-ACUGCA-----GCUCAAAACAAAGUUAUUACGGCCUAAGCUCGGGCCUAGGCUUUGUUUGCCUUUCUGGCUUAUGGCUAAAAAAAAAACCCACAUGACCAGAUCAGCACC ((--.....-...)).-----.....(((((((((..(((.(((((......)))))))))))))))))((...(((((.(((((.................))))))))))...))... ( -28.93) >consensus GCUGCUCAA_ACUGCAGUGCGGCUCAAAACAAAGUUAUUACGGCCUAAG____________CUUUGUUUGCCUUUCUGGCUCAUGGCAAA_____AAACCCACAUGGCCAGAUCGGCCCC (((((.............)))))...((((((((...........................))))))))(((..(((((.(((((.................))))))))))..)))... (-24.41 = -24.63 + 0.22)

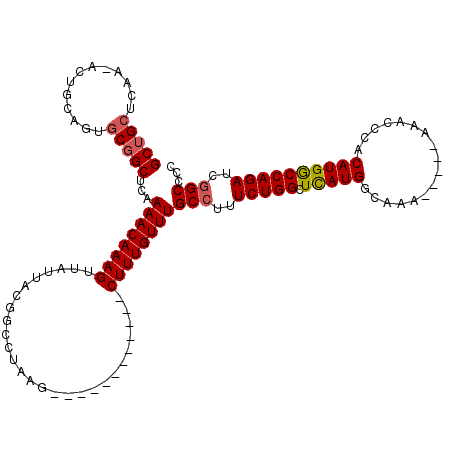
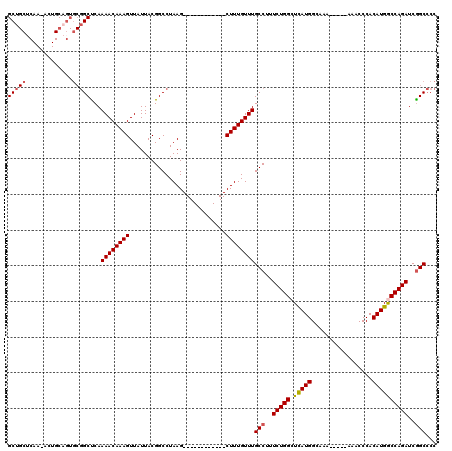
| Location | 10,748,268 – 10,748,371 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -22.89 |
| Energy contribution | -23.09 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10748268 103 + 23771897 CGGCCUAAGCUUUGUUUGCCUUUCUGGCUCAUGGCAAA-----AAACCCACAUGGCCAGAUCGGCCCCAGAUUUGCCCAAGCAUCAGUAUAUAUAUAUUUUUUCUUUU .(((..(((.((((...(((..((((((.((((.....-----.......))))))))))..)))..))))))))))............................... ( -24.80) >DroSec_CAF1 81768 98 + 1 CGGCCUAAGCUUUGUUUGCCUUUCUGGCUCAUGGCAAA-----AAACCCACAUGACCAGAUCGGCCCCAGAUUUGCCCCAGCAUAAGUAUCU----UUU-UUUUUACU .((..((((.((((...(((..(((((.(((((.....-----.......))))))))))..)))..))))))))..)).............----...-........ ( -22.90) >DroSim_CAF1 84394 99 + 1 CGGCCUAAGCUUUGUUUGCCUUUCUGGCUCAUGGCAAA-----AAACCCACAUGGCCAGAUCGGCCCCAGAUUUGCCCCAGCAUCAGUAUCU----UUUUUUUUUACU .((..((((.((((...(((..((((((.((((.....-----.......))))))))))..)))..))))))))..)).............----............ ( -25.00) >DroEre_CAF1 86675 94 + 1 CGGCCUAAGCUUUGUUUGCCUUUCUGGCUCAUGGCAAAA----AAACCCACAUGGCCAGAUCGGCCCCAGAUUUGCCCCAGCAUCAGUAUCU----UUU-UUU----- .((..((((.((((...(((..((((((.((((......----.......))))))))))..)))..))))))))..)).............----...-...----- ( -24.92) >DroYak_CAF1 97317 101 + 1 CGGCCUAAGCUUUGUUUGCCUUUCUGGCUCAUGGCAAAAAAAAAAACCCACAUGGCCAGAUCGGCCCCAGAUUUGCCCCAGCAUCAGUAUCU----UUU-UUU--UUU .((..((((.((((...(((..((((((.((((.................))))))))))..)))..))))))))..)).............----...-...--... ( -24.63) >consensus CGGCCUAAGCUUUGUUUGCCUUUCUGGCUCAUGGCAAA_____AAACCCACAUGGCCAGAUCGGCCCCAGAUUUGCCCCAGCAUCAGUAUCU____UUU_UUU_UACU .(((..(((.((((...(((..((((((.((((.................))))))))))..)))..))))))))))............................... (-22.89 = -23.09 + 0.20)

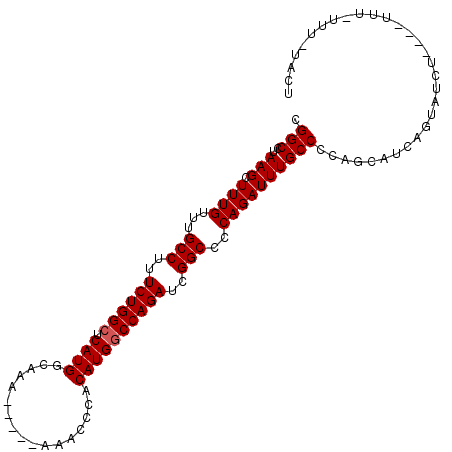

| Location | 10,748,268 – 10,748,371 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 90.43 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.14 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10748268 103 - 23771897 AAAAGAAAAAAUAUAUAUAUACUGAUGCUUGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU-----UUUGCCAUGAGCCAGAAAGGCAAACAAAGCUUAGGCCG ..........................((((((((.....((..(((..((((((((((.(.(...-----..).))))).))))))..)))...))..)))))))).. ( -31.30) >DroSec_CAF1 81768 98 - 1 AGUAAAAA-AAA----AGAUACUUAUGCUGGGGCAAAUCUGGGGCCGAUCUGGUCAUGUGGGUUU-----UUUGCCAUGAGCCAGAAAGGCAAACAAAGCUUAGGCCG ........-...----..........(((..(((.....((..(((..((((((((((.(.(...-----..).)))))).)))))..)))...))..)))..))).. ( -26.40) >DroSim_CAF1 84394 99 - 1 AGUAAAAAAAAA----AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU-----UUUGCCAUGAGCCAGAAAGGCAAACAAAGCUUAGGCCG ............----..........(((..(((.....((..(((..((((((((((.(.(...-----..).))))).))))))..)))...))..)))..))).. ( -28.50) >DroEre_CAF1 86675 94 - 1 -----AAA-AAA----AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU----UUUUGCCAUGAGCCAGAAAGGCAAACAAAGCUUAGGCCG -----...-...----..........(((..(((.....((..(((..((((((((((.(.(...----...).))))).))))))..)))...))..)))..))).. ( -28.70) >DroYak_CAF1 97317 101 - 1 AAA--AAA-AAA----AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUUUUUUUUUUGCCAUGAGCCAGAAAGGCAAACAAAGCUUAGGCCG ...--...-...----..........(((..(((.....((..(((..((((((((((..((........))..).))).))))))..)))...))..)))..))).. ( -28.60) >consensus AAAA_AAA_AAA____AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU_____UUUGCCAUGAGCCAGAAAGGCAAACAAAGCUUAGGCCG ..........................(((..(((.....((..(((..((((((((((..((........))..).))).))))))..)))...))..)))..))).. (-27.30 = -27.14 + -0.16)


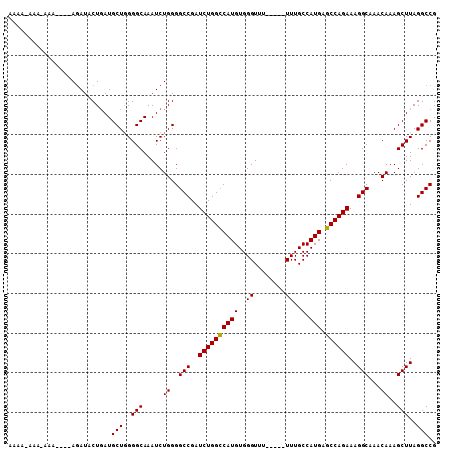
| Location | 10,748,296 – 10,748,411 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.69 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -26.56 |
| Energy contribution | -26.64 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10748296 115 - 23771897 CAUGUGGUUGGGGGCUAAUGCUGAGGCUAAAAACAGACAGAAAAGAAAAAAUAUAUAUAUACUGAUGCUUGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU-----UUUGCCAUGA ((((((((..(((((....)))..((((.....(((((((.....................))).(((....)))..)))).))))..))..))))))))(((..-----...))).... ( -32.40) >DroSec_CAF1 81796 110 - 1 CAUGUGGUUGGGGGCUAAUGCUGAGGCUAAAAACAGAAAGAGUAAAAA-AAA----AGAUACUUAUGCUGGGGCAAAUCUGGGGCCGAUCUGGUCAUGUGGGUUU-----UUUGCCAUGA (((((((..((((((....)))..((((.....((((..(((((....-...----...))))).(((....)))..)))).))))..)))..)))))))(((..-----...))).... ( -29.80) >DroSim_CAF1 84422 111 - 1 CAUGUGGUUGGGGGCUAAUGCUGAGGCUAAAAACAGAAAGAGUAAAAAAAAA----AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU-----UUUGCCAUGA ((((((((..(((((....)))..((((.....((((...((((........----...))))..(((....)))..)))).))))..))..))))))))(((..-----...))).... ( -33.00) >DroEre_CAF1 86703 104 - 1 CAUGUGGUUGGGGGCUAAUGCUGAGGCUAAAAACCGAA-------AAA-AAA----AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU----UUUUGCCAUGA ((((((((..((((((..((((..(((...........-------...-...----..........)))..)))).......))))..))..))))))))(((..----....))).... ( -28.43) >DroYak_CAF1 97345 113 - 1 CAUGUGGUUGGGGGCUAAUGCCGAGGCUAAAAACAGAAAGAAA--AAA-AAA----AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUUUUUUUUUUGCCAUGA ((((((((..(((((....)))..((((.....((((......--...-...----((...))..(((....)))..)))).))))..))..))))))))(((..........))).... ( -31.40) >consensus CAUGUGGUUGGGGGCUAAUGCUGAGGCUAAAAACAGAAAGAAAA_AAA_AAA____AGAUACUGAUGCUGGGGCAAAUCUGGGGCCGAUCUGGCCAUGUGGGUUU_____UUUGCCAUGA ((((((((..(((((....)))..((((.....((((...................((...))..(((....)))..)))).))))..))..))))))))(((..........))).... (-26.56 = -26.64 + 0.08)

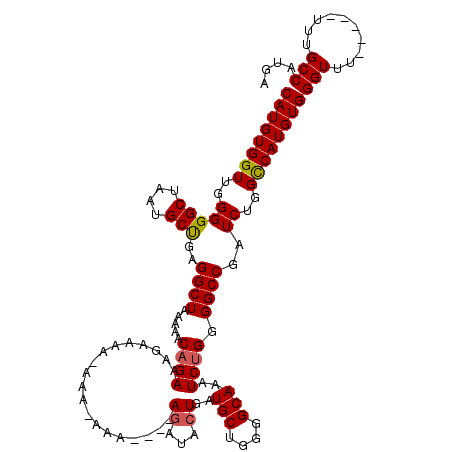
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:08 2006