| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,743,474 – 10,743,579 |
| Length | 105 |
| Max. P | 0.800842 |

| Location | 10,743,474 – 10,743,579 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.27 |
| Mean single sequence MFE | -26.07 |
| Consensus MFE | -19.00 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
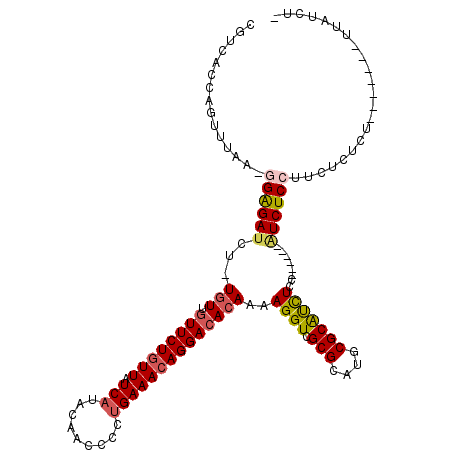
>3L_DroMel_CAF1 10743474 105 + 23771897 CGUCACCAGUUUAA-GGAGAUCU-UGUUGUUCUGUUAUCAUCCAACCCCCGAAACAGGACACAAAAGGUCGCGCAUGCGCAUCUCC------AUCUCAUUUUCUCU-------UUAUCUC ..............-((((((((-(..(((((((((.((...........)))))))))))...))))))(((....)))...)))------..............-------....... ( -24.40) >DroSec_CAF1 77005 111 + 1 CGUCACCAGUUUAA-GGAGAUCU-UGUUGUUCUGUUAUCAUACAACCCCUGAAACAGGACACAAAAGGUCGCGCAUGCGCAUCUCCAUCUCCAUCUCCUUUUCUCU-------UUAUCUC .......((...((-((((((.(-(((.((((((((.(((.........))))))))))))))).((((.(((....)))))))........))))))))..))..-------....... ( -27.50) >DroSim_CAF1 79624 111 + 1 CGUCACCAGUUUAA-GGAGAUCU-UGUUGUUCUGUUAUCAUACAACCCCUGAAACAGGACACAAAAGGUCGCGCAUGCGCAUCUCCAUCUCCAUCUCCUUUUCUCU-------UUAUCUC .......((...((-((((((.(-(((.((((((((.(((.........))))))))))))))).((((.(((....)))))))........))))))))..))..-------....... ( -27.50) >DroEre_CAF1 82038 104 + 1 CGUCACCAGUUUAA-GGAGAUCU-UGUUGUUCUGUUAUCAUACAACCCCUGAAACAGGACACAAAAGGUCGCGCAUGCGCAUCUCC------GUCUCCUUCUCUCU-------UUAUCU- ............((-((((((.(-(((.((((((((.(((.........))))))))))))))).((((.(((....)))))))..------))))))))......-------......- ( -27.20) >DroYak_CAF1 92447 104 + 1 CGUCACCAGUUUAA-GGAGAUCU-UGUUGUUCUGUUAUCAUACAACCCCUGAAACAGGACACAAAAGGUCGCGCAUGCGCAUUUCU------AUCUCCCUCUCUCA-------UUACCG- ..............-((((((((-(..(((((((((.(((.........))))))))))))...))))))(((....)))......------...)))........-------......- ( -24.60) >DroAna_CAF1 79813 107 + 1 CGUCAACAGUUCAAAGGGGAUAUAUCUCCUUCUAUUAUCAU-----CCCUGAAAAAGGACACAAAAGGUUGCGCAUGCGCGCCU---UCUCACUCUC--UCUCUCUGGGCGACUUAU--- .(((.........(((((((....)))))))..........-----(((.((.(.((((.....(((((.(((....)))))))---).....)).)--).).)).))).)))....--- ( -25.20) >consensus CGUCACCAGUUUAA_GGAGAUCU_UGUUGUUCUGUUAUCAUACAACCCCUGAAACAGGACACAAAAGGUCGCGCAUGCGCAUCUCC______AUCUCCUUCUCUCU_______UUAUCU_ ...............((((((...(((.((((((((.(((.........))))))))))))))..((((.(((....)))))))........))))))...................... (-19.00 = -19.50 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:04:02 2006