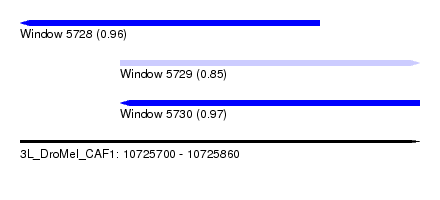
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,725,700 – 10,725,860 |
| Length | 160 |
| Max. P | 0.965560 |

| Location | 10,725,700 – 10,725,820 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -15.59 |
| Energy contribution | -16.15 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725700 120 - 23771897 GCAAAUGAUUAUUUCAUUUCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUCAGCCUUCCUCAGGAGCUUUCAUUUGCCCAUUGGCA ((((((((.............((..((((((((...(((((((....)))))))....))))))))..))...............(((((......)).))).))))))))......... ( -28.30) >DroSec_CAF1 59541 120 - 1 GCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUUGCCUGCCUCAGGAGCUUUCAUUUGGCCAUUGGCA ...(((((.....)))))(((((..((((((((...(((((((....)))))))....)))))))).........(((((.......((((...)))).......)))))....))))). ( -31.94) >DroSim_CAF1 61725 120 - 1 GCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUUGCCUGCCUCAGGAGCUUUCAUUUGGCCAUUGGCA ...(((((.....)))))(((((..((((((((...(((((((....)))))))....)))))))).........(((((.......((((...)))).......)))))....))))). ( -31.94) >DroYak_CAF1 74159 100 - 1 GCAAAUG------------AUUAUUUCAAGUUAUUAAAUUCCUUCUACGGUAUACACUUAGUUUGAAAUUCUUUACAAAUUAUGUAGCAUUCGCCAGGAGCUUUAA--------UGGCCA .....((------------(.....))).((((((((((((((....(((..((((..(((((((.((....)).)))))))))))....)))..))))).)))))--------)))).. ( -18.60) >consensus GCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAUUAUUUAGCCUGCCUCAGGAGCUUUCAUUUGGCCAUUGGCA ..................(((((..((((((((...(((((((....)))))))....))))))))..((((.......................))))...............))))). (-15.59 = -16.15 + 0.56)


| Location | 10,725,740 – 10,725,860 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -24.83 |
| Energy contribution | -24.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725740 120 + 23771897 AUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGAAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGCAAUUGUAAUGAGCACG ..............((((.(((..(((((((((....))))))))).))).))))(((((.((((((.....)))))).)))))..........(.((((((((....)))))))))... ( -34.40) >DroSec_CAF1 59581 120 + 1 AUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGCAAGAAAGACUUCUUGAAAUUGGAAUGAGUAGG ..((((...(((((.(((.(((..(((((((((....))))))))).))).)))))))).))))........((((((.(((.....((((((....))))))...)))))))))..... ( -29.30) >DroSim_CAF1 61765 120 + 1 AUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGAAACUGGAAUGAGUAGG ..............((((.(((..(((((((((....))))))))).))).))))(((((.((((((.....))).))))))))...........(((((((........)))))))... ( -27.10) >consensus AUUUGUAAAGGAUUUCAAACUAAAUGAAUUCCGUAGACGGAAUUUAAUAGCUUGAGUUCGGCAAUGAAAUAAUCAUUUGCGAGCAUGAAAGAAAGACUUAUUGAAAUUGGAAUGAGUAGG .......(((..((((........(((((((((....))))))))).........(((((..(((((.....)))))..)))))......))))..)))..................... (-24.83 = -24.83 + -0.00)



| Location | 10,725,740 – 10,725,860 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -24.17 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.53 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725740 120 - 23771897 CGUGCUCAUUACAAUUGCAAUAAGUCUUUCUUUCAUGCUCGCAAAUGAUUAUUUCAUUUCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAU .(((............(((..(((.....)))...)))(((.((((((.....)))))).)))..((((((((...(((((((....)))))))....)))))))).......))).... ( -26.20) >DroSec_CAF1 59581 120 - 1 CCUACUCAUUCCAAUUUCAAGAAGUCUUUCUUGCAUGCUCGCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAU .................((((((....)))))).....(((..(((((.....)))))..)))..((((((((...(((((((....)))))))....)))))))).............. ( -24.70) >DroSim_CAF1 61765 120 - 1 CCUACUCAUUCCAGUUUCAAUAAGUCUUUCUUUCAUGCUCGCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAU ......................................(((..(((((.....)))))..)))..((((((((...(((((((....)))))))....)))))))).............. ( -21.60) >consensus CCUACUCAUUCCAAUUUCAAUAAGUCUUUCUUUCAUGCUCGCAAAUGAUUAUUUCAUUGCCGAACUCAAGCUAUUAAAUUCCGUCUACGGAAUUCAUUUAGUUUGAAAUCCUUUACAAAU ......................................(((..(((((.....)))))..)))..((((((((...(((((((....)))))))....)))))))).............. (-21.53 = -21.53 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:49 2006