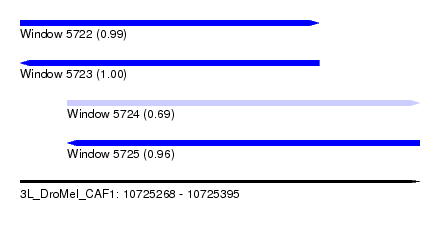
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,725,268 – 10,725,395 |
| Length | 127 |
| Max. P | 0.999923 |

| Location | 10,725,268 – 10,725,363 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -22.15 |
| Consensus MFE | -19.80 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
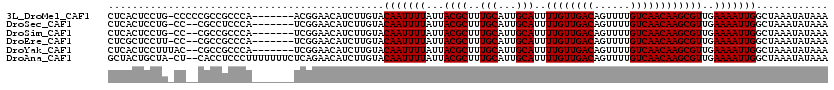
>3L_DroMel_CAF1 10725268 95 + 23771897 --------------------GGAUGGGU-----ACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG --------------------...(((.(-----(.((((....)))).)).))).......(((((((((((((....)))))))....(((...)))))))))................ ( -21.30) >DroSec_CAF1 59131 95 + 1 --------------------GGAUGGGU-----ACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG --------------------...(((.(-----(.((((....)))).)).))).......(((((((((((((....)))))))....(((...)))))))))................ ( -21.30) >DroSim_CAF1 61303 95 + 1 --------------------GGAUGGGU-----ACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG --------------------...(((.(-----(.((((....)))).)).))).......(((((((((((((....)))))))....(((...)))))))))................ ( -21.30) >DroEre_CAF1 63941 95 + 1 --------------------GGAUGGGU-----ACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG --------------------...(((.(-----(.((((....)))).)).))).......(((((((((((((....)))))))....(((...)))))))))................ ( -21.30) >DroYak_CAF1 73697 95 + 1 --------------------GGAUGGGU-----ACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG --------------------...(((.(-----(.((((....)))).)).))).......(((((((((((((....)))))))....(((...)))))))))................ ( -21.30) >DroAna_CAF1 61190 120 + 1 AUGGACGGGGGGAGAUGGAUGGAUGGGUACAGUACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG .((.((((..(((.(((((((.(((.........))).))))))).)))..))((((((..(((((((((((((....)))))))....(((...)))))))))...)))))))).)).. ( -26.40) >consensus ____________________GGAUGGGU_____ACAUAUAUUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAG ....................................................(((((((..(((((((((((((....)))))))....(((...)))))))))...)))))))...... (-19.80 = -19.80 + 0.00)

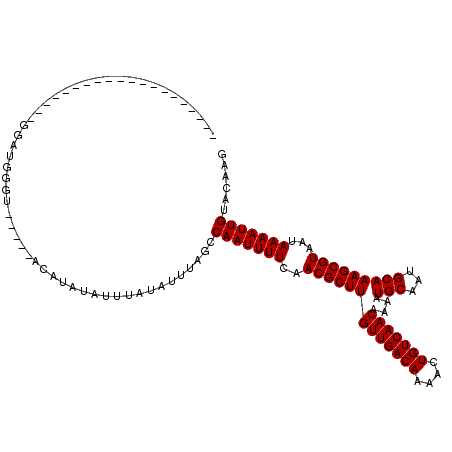
| Location | 10,725,268 – 10,725,363 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.58 |
| SVM RNA-class probability | 0.999923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725268 95 - 23771897 CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGU-----ACCCAUCC-------------------- ......(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))...................-----........-------------------- ( -21.30) >DroSec_CAF1 59131 95 - 1 CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGU-----ACCCAUCC-------------------- ......(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))...................-----........-------------------- ( -21.30) >DroSim_CAF1 61303 95 - 1 CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGU-----ACCCAUCC-------------------- ......(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))...................-----........-------------------- ( -21.30) >DroEre_CAF1 63941 95 - 1 CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGU-----ACCCAUCC-------------------- ......(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))...................-----........-------------------- ( -21.30) >DroYak_CAF1 73697 95 - 1 CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGU-----ACCCAUCC-------------------- ......(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))...................-----........-------------------- ( -21.30) >DroAna_CAF1 61190 120 - 1 CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGUACUGUACCCAUCCAUCCAUCUCCCCCCGUCCAU ......(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))((...(((((....)))))...))............................ ( -21.90) >consensus CUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAAUAUAUGU_____ACCCAUCC____________________ ......(((((((...((((..(((...)))..((((((((......))))))))))))..))))))).................................................... (-21.30 = -21.30 + 0.00)

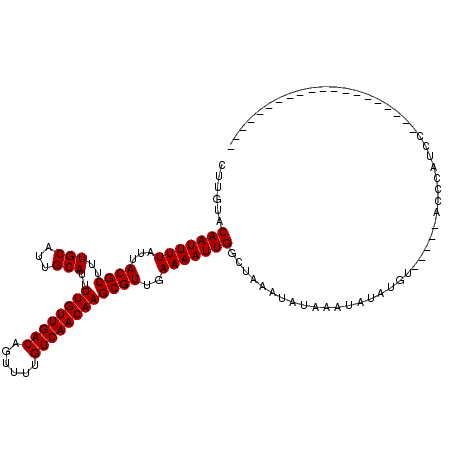
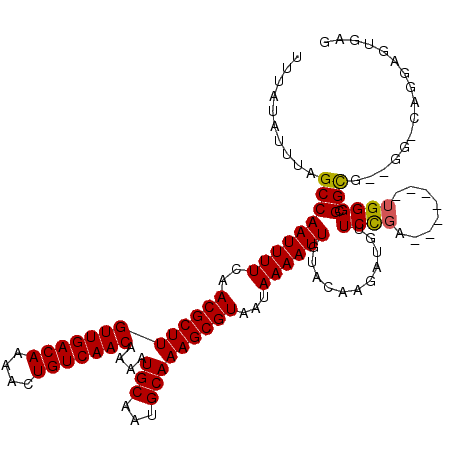
| Location | 10,725,283 – 10,725,395 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -23.46 |
| Energy contribution | -23.35 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725283 112 + 23771897 UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCCGU-------UGGGCGGCGGGGG-CAGGAGUGAG ....(((((.(((((((((..(((((((((((((....)))))))....(((...)))))))))...))))))...........(((((-------(....))))))))-)..))))).. ( -29.90) >DroSec_CAF1 59146 110 + 1 UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCCGA-------UGGGAGGCG--GG-CAGGAGUGAG ....(((((.(((((((((..(((((((((((((....)))))))....(((...)))))))))...))))))........((((((..-------..))).)))--))-)..))))).. ( -26.60) >DroSim_CAF1 61318 110 + 1 UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCCGA-------UGGGCGGCG--GG-CAGGAGUGAG ....(((((.(((((((((..(((((((((((((....)))))))....(((...)))))))))...))))))........((((((..-------..)).))))--))-)..))))).. ( -25.60) >DroEre_CAF1 63956 110 + 1 UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCCGA-------UGGGCGGCG--GG-AAGGAGCGAG ............(((((((..(((((((((((((....)))))))....(((...)))))))))...))))))).......((((((..-------(.(....).--).-..)))))).. ( -26.60) >DroYak_CAF1 73712 111 + 1 UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCCGA-------UGGGCGGCG--GUAAAGGAGUGAG (((((.....(((((((((..(((((((((((((....)))))))....(((...)))))))))...)))))).........((((...-------.))))))).--)))))........ ( -25.10) >DroAna_CAF1 61230 117 + 1 UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCUGAGAAAAAAAGGGAGGUG--AG-UAGCAGUAGC ..........(((((((((..(((((((((((((....)))))))....(((...)))))))))...)))))))(((...((.((((..........)))).)).--.)-))))...... ( -23.50) >consensus UUUAUAUUUAGCCAAUUUUCAACGCUUGUUGACAAAACUGUCAACAAAAUGCAAUGCAAAGCGUAAUAAAAUUGUACAAGAUGUUCCGA_______UGGGCGGCG__GG_CAGGAGUGAG ..........(((((((((..(((((((((((((....)))))))....(((...)))))))))...))))))...........((((........)))).)))................ (-23.46 = -23.35 + -0.11)


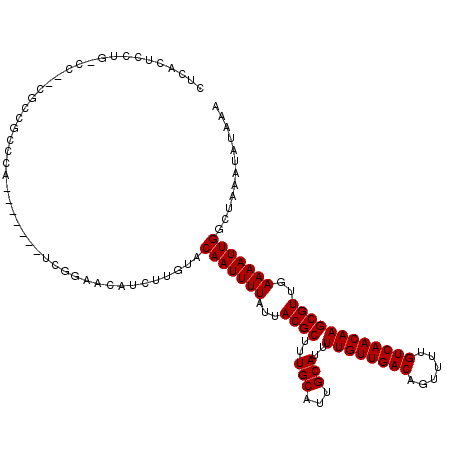
| Location | 10,725,283 – 10,725,395 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.71 |
| Mean single sequence MFE | -23.92 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.963660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725283 112 - 23771897 CUCACUCCUG-CCCCCGCCGCCCA-------ACGGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA .........(-(..(((.......-------.)))...........(((((((...((((..(((...)))..((((((((......))))))))))))..))))))))).......... ( -24.70) >DroSec_CAF1 59146 110 - 1 CUCACUCCUG-CC--CGCCUCCCA-------UCGGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA ..........-..--.((((((..-------..)))...........((((((...((((..(((...)))..((((((((......))))))))))))..))))))))).......... ( -24.10) >DroSim_CAF1 61318 110 - 1 CUCACUCCUG-CC--CGCCGCCCA-------UCGGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA .........(-((--((.......-------.)))...........(((((((...((((..(((...)))..((((((((......))))))))))))..))))))))).......... ( -24.10) >DroEre_CAF1 63956 110 - 1 CUCGCUCCUU-CC--CGCCGCCCA-------UCGGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA ...((.....-.(--((.......-------.)))...........(((((((...((((..(((...)))..((((((((......))))))))))))..))))))))).......... ( -24.80) >DroYak_CAF1 73712 111 - 1 CUCACUCCUUUAC--CGCCGCCCA-------UCGGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA ............(--((.......-------.)))...........(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))............ ( -23.50) >DroAna_CAF1 61230 117 - 1 GCUACUGCUA-CU--CACCUCCCUUUUUUUCUCAGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA ((....))..-..--...............................(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))............ ( -22.30) >consensus CUCACUCCUG_CC__CGCCGCCCA_______UCGGAACAUCUUGUACAAUUUUAUUACGCUUUGCAUUGCAUUUUGUUGACAGUUUUGUCAACAAGCGUUGAAAAUUGGCUAAAUAUAAA ..............................................(((((((...((((..(((...)))..((((((((......))))))))))))..)))))))............ (-21.30 = -21.30 + 0.00)
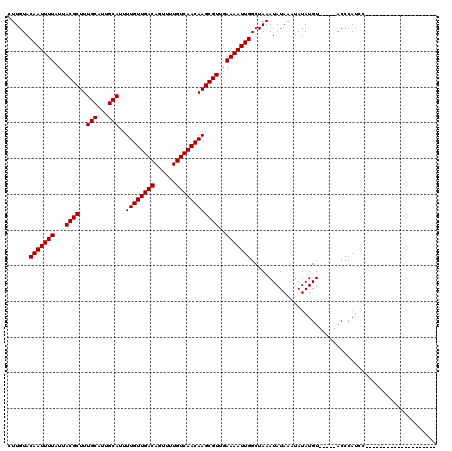
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:45 2006