
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,724,971 – 10,725,259 |
| Length | 288 |
| Max. P | 0.938175 |

| Location | 10,724,971 – 10,725,085 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -31.95 |
| Energy contribution | -31.95 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10724971 114 - 23771897 GGUUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACC---ACCAG (((.((.(---(((....))))...(((.....((((((((((..(((.....))).....)))))))))).......((((((..(((...))).)))))).)))....))---))).. ( -36.60) >DroSec_CAF1 58859 114 - 1 GGAUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAACAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACC---ACCAG ((.(((.(---(((....))))...(((..(((....))).....((.(((((((((......))))))))).))...((((((..(((...))).)))))).)))....))---))).. ( -35.20) >DroSim_CAF1 61031 114 - 1 GGUUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACC---ACCAG (((.((.(---(((....))))...(((.....((((((((((..(((.....))).....)))))))))).......((((((..(((...))).)))))).)))....))---))).. ( -36.60) >DroEre_CAF1 63633 114 - 1 GGAUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACC---ACCAG ((.(((.(---(((....))))...(((.....((((((((((..(((.....))).....)))))))))).......((((((..(((...))).)))))).)))....))---))).. ( -36.60) >DroYak_CAF1 73374 117 - 1 GGCUGGGGCUGCAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACC---ACCAG ((.(((((((((((....))))..))))..((.((((((((((..(((.....))).....)))))))))).......((((((..(((...))).))))))..))....))---))).. ( -38.60) >DroAna_CAF1 60884 117 - 1 AGGUGGGG---UAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACCACCACCAG .(((((((---(((((((((((...(.....).((((((((((..(((.....))).....))))))))))..)))))((((((..(((...))).))))))))).)).))).))))).. ( -42.40) >consensus GGAUGGGG___CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUGGCAAAGUGCAAAGUAGUUGCCAAGGCAUCACC___ACCAG ...............(((((((((.((((.(((....))).....((.(((((((((......))))))))).))...))))...))))......(.(((....))).)......))))) (-31.95 = -31.95 + -0.00)



| Location | 10,725,008 – 10,725,121 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.22 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725008 113 - 23771897 AGGGUACUUUGUAC-AGGGUACGA--AGAAGGGGAAU-GGGGUUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG .(((((((((((((-...))))))--)..........-..(..((..(---(((....)))).))..).....((((((((((..(((.....))).....))))))))))..)))))). ( -32.90) >DroSec_CAF1 58896 90 - 1 AGGGUA--------------------------GGGAU-GGGGAUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAACAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG .((.(.--------------------------.(.((-.((((((...---)))))).)).)..).))..(((....))).....((.(((((((((......))))))))).))..... ( -24.90) >DroSim_CAF1 61068 90 - 1 AGGAUA--------------------------GGGAU-GGGGUUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG .(((((--------------------------.....-..(..((..(---(((....)))).))..).....((((((((((..(((.....))).....))))))))))...))))). ( -26.80) >DroEre_CAF1 63670 115 - 1 AGGGUACUUUGUAC-AGGGUAUGGGUGUACGAGGGAU-GAGGAUGGGG---CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG .((((((((.....-...((.((.((((((..(((((-(.........---)))))).))))))...)).)).((((((((((..(((.....))).....)))))))))))))))))). ( -39.50) >DroYak_CAF1 73411 119 - 1 AGGGUACUUUAUACCAGGGUAUGGGUGGUGGAGGGAU-GAGGCUGGGGCUGCAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG .((((((((.........((.(((.(((((..(((((-(.(((....))).))))))..))).)).))).)).((((((((((..(((.....))).....)))))))))))))))))). ( -43.10) >DroAna_CAF1 60924 93 - 1 GGGAUA------------------------GAGGGGUCCUAGGUGGGG---UAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG .(((((------------------------((..((..(((((.....---...)))))..).)..)).....((((((((((..(((.....))).....))))))))))...))))). ( -28.20) >consensus AGGGUA________________________GAGGGAU_GGGGAUGGGG___CAUCCUGGUGCACAGUCAAGCGAAUUCGCAGCAAGCAUGACUUGUAAAUAGCUGCGAGUUAAGUAUCUG ............................................((((.....))))(((((........(((....)))........(((((((((......))))))))).))))).. (-19.32 = -19.32 + -0.00)
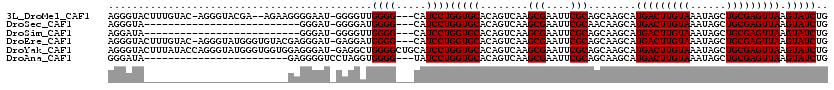

| Location | 10,725,121 – 10,725,219 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -23.30 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725121 98 + 23771897 AUAGCUAUCC----------------------CCCCUCGCCAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUU ..........----------------------...((((((.....)))((((((...))))))(((((..(((..((((((((........))))))))...))))))))....))).. ( -24.20) >DroSec_CAF1 58986 96 + 1 AUAGCCAUCC----------------------CCC--CGCCAUUUGGGCACAGAUGAAAUCAGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUU .....((((.----------------------.((--((.....))))....))))...((..((((((..(((..((((((((........))))))))...)))))))))...))... ( -20.50) >DroSim_CAF1 61158 96 + 1 AUAGCCAUCC----------------------CCC--CGCCAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUU ..........----------------------...--(((.......((((((((...))))))))...(((((..((((((((........))))))))...))))).)))........ ( -22.40) >DroEre_CAF1 63785 107 + 1 GUAACUAUGC-----------CCCUCCCCGCCCCG--CGCCAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGUUGCGAACGAGUU ..........-----------..(((..(((..((--(((.......((((((((...)))))))).....))...((((((((........))))))))....)))..)))...))).. ( -25.90) >DroYak_CAF1 73530 118 + 1 GUAGCUAUUCCACCUCCCUCCCUCUCCCCACCCCA--CGCCUUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUU .................(((..((...........--.(((.....)))((((((...))))))(((((..(((..((((((((........))))))))...))))))))))..))).. ( -24.40) >DroAna_CAF1 61017 93 + 1 ACCCUGAUCA----------------------UCG--CGCU-UUUAGG--CAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAAUGCGAUGCGAACGAGUU .......(((----------------------((.--.(((-....))--).)))))..((..((((((..(((..((((((((........))))))))...)))))))))...))... ( -22.40) >consensus AUAGCUAUCC______________________CCC__CGCCAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUU .....................................(((.......((((((((...))))))))...(((((..((((((((........))))))))...))))).)))........ (-19.20 = -19.87 + 0.67)

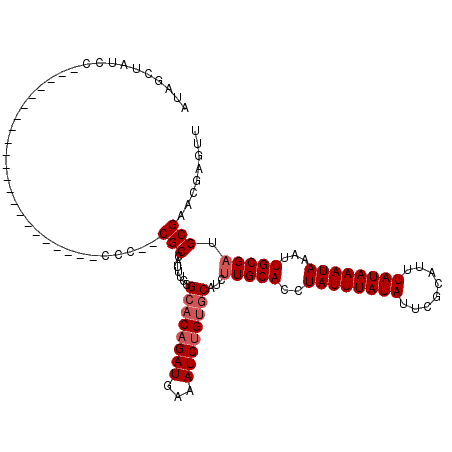
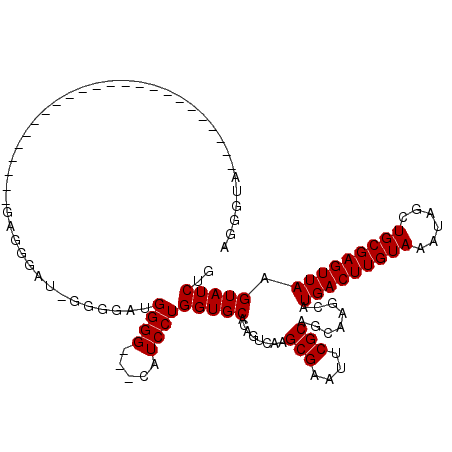
| Location | 10,725,121 – 10,725,219 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.04 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -19.53 |
| Energy contribution | -20.03 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725121 98 - 23771897 AACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUGGCGAGGGG----------------------GGAUAGCUAU ..(((.(((((...(((.((((((((((........))))))))))))).....((((((((...)))))))).......))))).))----------------------)......... ( -31.10) >DroSec_CAF1 58986 96 - 1 AACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACUGAUUUCAUCUGUGCCCAAAUGGCG--GGG----------------------GGAUGGCUAU ...((((((((((((((.((((((((((........)))))))))))))..)))))...........((.((((.....))))--.))----------------------)))))).... ( -26.50) >DroSim_CAF1 61158 96 - 1 AACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUGGCG--GGG----------------------GGAUGGCUAU ..(((.(((((...(((.((((((((((........))))))))))))).....((((((((...)))))))).......)))--)))----------------------))........ ( -27.20) >DroEre_CAF1 63785 107 - 1 AACUCGUUCGCAACGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUGGCG--CGGGGCGGGGAGGG-----------GCAUAGUUAC ..(((.(((((..(((.(((((((((((........)))))))))))((.....((((((((...)))))))).......)))--))..))))))))..-----------.......... ( -34.50) >DroYak_CAF1 73530 118 - 1 AACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAAGGCG--UGGGGUGGGGAGAGGGAGGGAGGUGGAAUAGCUAC ..(((.(((((((((((.((((((((((........)))))))))))))..))))).....(..((((..((((.....))))--..).)))..).))).)))....((((.....)))) ( -35.50) >DroAna_CAF1 61017 93 - 1 AACUCGUUCGCAUCGCAUUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUG--CCUAAA-AGCG--CGA----------------------UGAUCAGGGU .((((....(((((((((((.(((((((........)))))))))))))..)))))...((..(((((((--(.....-.)))--.))----------------------))))).)))) ( -25.10) >consensus AACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUGGCG__GGG______________________GGAUAGCUAU ........(((...(((.((((((((((........))))))))))))).....((((((((...)))))))).......)))..................................... (-19.53 = -20.03 + 0.50)

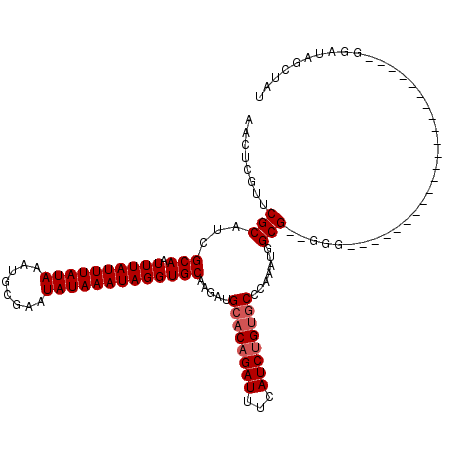

| Location | 10,725,139 – 10,725,259 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -26.04 |
| Energy contribution | -26.87 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725139 120 + 23771897 CAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAGUGCGACAA .......((((((((...))))))))...((((((.((((((((........)))))))).((((((((((((........))).))))))))).................))))))... ( -30.10) >DroSec_CAF1 59002 120 + 1 CAUUUGGGCACAGAUGAAAUCAGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAGUGCGACAA ((((((....))))))......((((.....)))).(((((((..(((((((((....)))).)))))(((.((((.((....))))))))))))))))....(((((...))))).... ( -27.30) >DroSim_CAF1 61174 120 + 1 CAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAGUGCGACAA .......((((((((...))))))))...((((((.((((((((........)))))))).((((((((((((........))).))))))))).................))))))... ( -30.10) >DroEre_CAF1 63812 120 + 1 CAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGUUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAGUGCGACAA .......((((((((...)))))))).....(((..((((((((........))))))))...)))((((((((....(((((((....)))))))...)))(((....))).))))).. ( -30.10) >DroYak_CAF1 73568 120 + 1 CUUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAGUGCGACAA .......((((((((...))))))))...((((((.((((((((........)))))))).((((((((((((........))).))))))))).................))))))... ( -30.10) >DroAna_CAF1 61033 117 + 1 U-UUUAGG--CAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAAUGCGAUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAUUGCGACAA .-....((--(((......((..((((((..(((..((((((((........))))))))...)))))))))...))....)))))((((((((................)))))))).. ( -27.59) >consensus CAUUUGGGCACAGAUGAAAUCUGUGCAUCUUGCACCUAUUUAUAUUCGCAUUUAUAAAUAAAUUGCGAUGCGAACGAGUUGUUGCCGUUGUAAUAAAUAUUCCUGCACACAGUGCGACAA .......((((((((...))))))))...((((((..........(((((((((....)))).)))))((((......(((((((....))))))).......))))....))))))... (-26.04 = -26.87 + 0.83)



| Location | 10,725,139 – 10,725,259 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.28 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -24.13 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10725139 120 - 23771897 UUGUCGCACUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUG .....((((...))))..................(((............((((((((.((((((((((........)))))))))))))..)))))((((((...)))))))))...... ( -28.10) >DroSec_CAF1 59002 120 - 1 UUGUCGCACUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACUGAUUUCAUCUGUGCCCAAAUG (((..((((.(((..(((................(....).........((((((((.((((((((((........)))))))))))))..))))).)))....)))..)))).)))... ( -27.00) >DroSim_CAF1 61174 120 - 1 UUGUCGCACUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUG .....((((...))))..................(((............((((((((.((((((((((........)))))))))))))..)))))((((((...)))))))))...... ( -28.10) >DroEre_CAF1 63812 120 - 1 UUGUCGCACUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAACGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUG ..(((((.(((....)))................(....).........))...(((.((((((((((........)))))))))))))..)))((((((((...))))))))....... ( -27.00) >DroYak_CAF1 73568 120 - 1 UUGUCGCACUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAAG .....((((...))))..................(((............((((((((.((((((((((........)))))))))))))..)))))((((((...)))))))))...... ( -28.10) >DroAna_CAF1 61033 117 - 1 UUGUCGCAAUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAUCGCAUUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUG--CCUAAA-A .....(.(((.((((((....((((((((......(((((.....))).)).((((((((.......)))))))).....)))))))).....)))))).))).).....--......-. ( -25.80) >consensus UUGUCGCACUGUGUGCAGGAAUAUUUAUUACAACGGCAACAACUCGUUCGCAUCGCAAUUUAUUUAUAAAUGCGAAUAUAAAUAGGUGCAAGAUGCACAGAUUUCAUCUGUGCCCAAAUG .....((((...))))..................(((............((((((((.((((((((((........)))))))))))))..)))))((((((...)))))))))...... (-24.13 = -24.97 + 0.83)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:41 2006