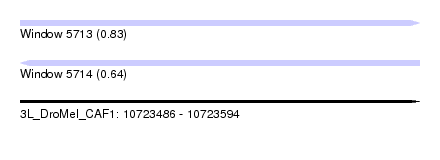
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,723,486 – 10,723,594 |
| Length | 108 |
| Max. P | 0.827332 |

| Location | 10,723,486 – 10,723,594 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -23.54 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10723486 108 + 23771897 UCUAAAAAUGUGACAAAAAAUGAUUUCGUUAAUGUCAGCAGAAAAUAAGUUUGCCCAAAGUCCAAG-CCUUUGGGAAACUUUUUGCUUCGUUUCAUUUUU-UUUUUUUAG .(((((((..(((((...((((....))))..)))))((((((....(((((.(((((((......-.))))))))))))))))))..............-..))))))) ( -26.10) >DroSec_CAF1 57438 108 + 1 UCUAC-AAUGUGACGAAAAAUGAUUUCGUUAAUGUCAGCAGAAAAUAAGUUGGCCCAAAGUCCAAG-CCUUUGGGAAACUUUUUGCUUCGUUUCAGUUUUUUUUUUUUAG ...((-(...((((((((.....)))))))).))).(((((((....((((..(((((((......-.))))))).)))))))))))....................... ( -24.30) >DroSim_CAF1 59603 104 + 1 UCUAC-AAUGUGACGAAAAAUGAUUUCGUUAAUGCCAGCAGAAAAUAAGUUGGCCCAAAGUCCAAG-CCUUUGGGAAACUUUUUGCUUCGUUUCAGUUUU----UUUUAG .....-..((.((((((.((((....)))).......((((((....((((..(((((((......-.))))))).)))))))))))))))).)).....----...... ( -24.10) >DroEre_CAF1 62083 102 + 1 UCUAC-AAUGUGACGAUAAAUGAUUUCGUUAAUGUCAGCACAAAAUAAGUUGGCCUAAAGUCCAAGUCCAUUGGGAAACUUUUUGCUUCGUUUCAGUUU-------UUAG ....(-((((.(((....((((....))))...((((((.........))))))...........)))))))).(((((..........))))).....-------.... ( -18.20) >DroYak_CAF1 71634 102 + 1 UCUAC-AAUGUGACGAAAAAUGAUUUCGUUAAUGUCAGCAGAAAAUAAGUUUGCCCAAAGUCCAAG-CCUUUGGGAAACUUUUUGCUUCGUUUCAGUUU------UUUAG ...((-(...((((((((.....)))))))).))).(((((((....(((((.(((((((......-.)))))))))))))))))))............------..... ( -25.00) >consensus UCUAC_AAUGUGACGAAAAAUGAUUUCGUUAAUGUCAGCAGAAAAUAAGUUGGCCCAAAGUCCAAG_CCUUUGGGAAACUUUUUGCUUCGUUUCAGUUUU____UUUUAG ........((.(((((..((((....))))......(((((((....((((..(((((((........))))))).)))))))))))))))).))............... (-19.84 = -20.28 + 0.44)

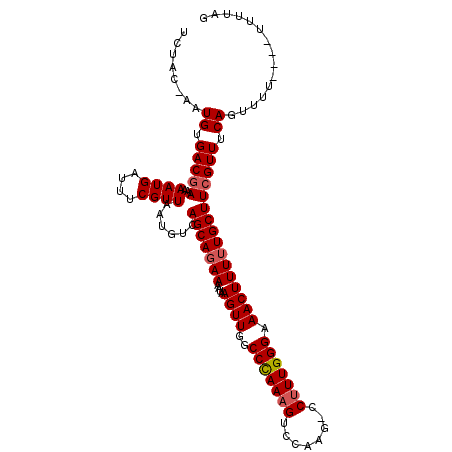
| Location | 10,723,486 – 10,723,594 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.16 |
| Mean single sequence MFE | -21.20 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.84 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10723486 108 - 23771897 CUAAAAAAA-AAAAAUGAAACGAAGCAAAAAGUUUCCCAAAGG-CUUGGACUUUGGGCAAACUUAUUUUCUGCUGACAUUAACGAAAUCAUUUUUUGUCACAUUUUUAGA (((((((.(-((((((((..((.((((..(((((((((((((.-......))))))).))))))......))))........))...)))))))))......))))))). ( -24.20) >DroSec_CAF1 57438 108 - 1 CUAAAAAAAAAAAACUGAAACGAAGCAAAAAGUUUCCCAAAGG-CUUGGACUUUGGGCCAACUUAUUUUCUGCUGACAUUAACGAAAUCAUUUUUCGUCACAUU-GUAGA .............................(((((.(((((((.-......)))))))..)))))....(((((........((((((.....))))))......-))))) ( -20.84) >DroSim_CAF1 59603 104 - 1 CUAAAA----AAAACUGAAACGAAGCAAAAAGUUUCCCAAAGG-CUUGGACUUUGGGCCAACUUAUUUUCUGCUGGCAUUAACGAAAUCAUUUUUCGUCACAUU-GUAGA ......----...................(((((.(((((((.-......)))))))..)))))....(((((........((((((.....))))))......-))))) ( -20.84) >DroEre_CAF1 62083 102 - 1 CUAA-------AAACUGAAACGAAGCAAAAAGUUUCCCAAUGGACUUGGACUUUAGGCCAACUUAUUUUGUGCUGACAUUAACGAAAUCAUUUAUCGUCACAUU-GUAGA ....-------...(((....(((((.....))))).((((((((((((.(.....)))))...(((((((..........)))))))........))).))))-)))). ( -16.80) >DroYak_CAF1 71634 102 - 1 CUAAA------AAACUGAAACGAAGCAAAAAGUUUCCCAAAGG-CUUGGACUUUGGGCAAACUUAUUUUCUGCUGACAUUAACGAAAUCAUUUUUCGUCACAUU-GUAGA .....------..................(((((((((((((.-......))))))).))))))....(((((........((((((.....))))))......-))))) ( -23.34) >consensus CUAAAA____AAAACUGAAACGAAGCAAAAAGUUUCCCAAAGG_CUUGGACUUUGGGCCAACUUAUUUUCUGCUGACAUUAACGAAAUCAUUUUUCGUCACAUU_GUAGA (((....................((((.(((((..(((((((........))))))).......))))).)))).......((((((.....))))))........))). (-16.40 = -16.84 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:34 2006