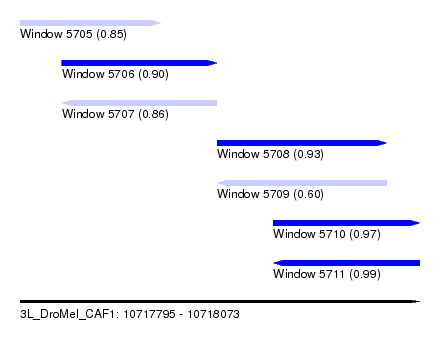
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,717,795 – 10,718,073 |
| Length | 278 |
| Max. P | 0.987488 |

| Location | 10,717,795 – 10,717,893 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.72 |
| Mean single sequence MFE | -18.18 |
| Consensus MFE | -9.70 |
| Energy contribution | -9.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717795 98 + 23771897 UCGGACUUCCCUCGGUAACCCCCCGUGAUU----------CCCCCCUCAACCGCCUUGUGUUUCCUCUCAUUUGGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUC ..((.((.(((.(((.......))).)...----------............(((..(((........)))..))).......)).))))..(((....)))...... ( -18.50) >DroSec_CAF1 51797 97 + 1 UCGGACUCCCCCCGGGAAUCCCCCGUGAUU----------CC-CCCUCAACCGCCUUGUGUUUCCUCUCAUCUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUC ..((.........(((((((......))))----------))-)............................((((((....))))))))..(((....)))...... ( -22.70) >DroSim_CAF1 53661 97 + 1 UUGGACUCCCCUCG-GAAUCCCCCGUGAUU----------CCCCCCUCAACCGCCUUGUGUUUCCUCUCAUCUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUC .(((.........(-(((((......))))----------))..............................((((((....))))))))).(((....)))...... ( -19.70) >DroYak_CAF1 65788 97 + 1 ACUGAUUCCCCACCG-AAUC----------CCCUUUGGAUCCCCCCCCAUCCGCCUUUUGUUUCCUCUCAUUUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUC ...(((((......)-))))----------......((((........))))....................((((((....))))))....(((....)))...... ( -17.00) >DroAna_CAF1 54072 82 + 1 GCUGACCCCCUCCU-CAACCCCU-------------------CUACCCAAUGGCUCUUUAU------CCAUUUGGCCAUAAUUGGCAGACACGCGUCUUCGCAUUUUC ..............-.......(-------------------((..(.(((((........------))))).)((((....)))))))...(((....)))...... ( -13.00) >consensus UCGGACUCCCCCCGGGAAUCCCCCGUGAUU__________CCCCCCUCAACCGCCUUGUGUUUCCUCUCAUUUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUC ..........................................................................((((....))))......(((....)))...... ( -9.70 = -9.70 + 0.00)

| Location | 10,717,824 – 10,717,932 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -35.45 |
| Consensus MFE | -28.61 |
| Energy contribution | -29.00 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717824 108 + 23771897 U-----------CCCCCCUCAACCGCCUUGUGUUUCCUCUCAUUUGGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUCGCUUGGCCGUGUAAAUUUGCGGAAUGCCUUUGAGGGGU-U .-----------..((((((((..(((..(((........)))..))).......((((((((.(((............))).))))((((......))))...)))).)))))))).-. ( -38.00) >DroSec_CAF1 51826 107 + 1 U-----------CC-CCCUCAACCGCCUUGUGUUUCCUCUCAUCUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUCGCUUGGCCGUGUAAAUUUGCGGAAUGCCUUUGAGGGGU-U .-----------.(-(((((((..((.((.(((.......(((..(((((....)))))((((.(((............))).)))).))).......))).)).))..)))))))).-. ( -36.74) >DroSim_CAF1 53689 108 + 1 U-----------CCCCCCUCAACCGCCUUGUGUUUCCUCUCAUCUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUCGCUUGGCCGUGUAAAUUUGCGGAAUGCCUUUGAGGGGU-U .-----------..((((((((..((.((.(((.......(((..(((((....)))))((((.(((............))).)))).))).......))).)).))..)))))))).-. ( -36.74) >DroEre_CAF1 56243 119 + 1 UCCCCUUCGGAUUCCCCCUCAUCCGCCUUGUGUUUCCUCUCAUUUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUCGCUUGGCCGUGUAAAUUUGCGGAAUGCCUUUGAGGGGU-U (((.....)))...(((((((...((.((.(((.......(((..(((((....)))))((((.(((............))).)))).))).......))).)).))...))))))).-. ( -36.84) >DroYak_CAF1 65807 117 + 1 --CCCUUUGGAUCCCCCCCCAUCCGCCUUUUGUUUCCUCUCAUUUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUCGCUUGGUCGUGUAAAUUUGCGGAAUGCCUUUGAGGGGU-U --((((((((........)))(((((..................((((((....))))))..(((((.((..((......))..)).)))))......)))))........)))))..-. ( -33.70) >DroAna_CAF1 54094 100 + 1 --------------CUACCCAAUGGCUCUUUAU------CCAUUUGGCCAUAAUUGGCAGACACGCGUCUUCGCAUUUUCGCUUGGCCGUGUAAAUUUGCGGAAUGCCUCUGAGGAGGCA --------------....((.((((((......------......)))))).....(((((.(((((.((..((......))..)).)))))...)))))))..((((((....)))))) ( -30.70) >consensus U___________CCCCCCUCAACCGCCUUGUGUUUCCUCUCAUUUUGCCAUAAUUGGCAGCCACGCGUCUUCGCAUUUUCGCUUGGCCGUGUAAAUUUGCGGAAUGCCUUUGAGGGGU_U ..............(((((((.........................((((....))))......((((.((((((..((..(........)..))..))))))))))...)))))))... (-28.61 = -29.00 + 0.39)

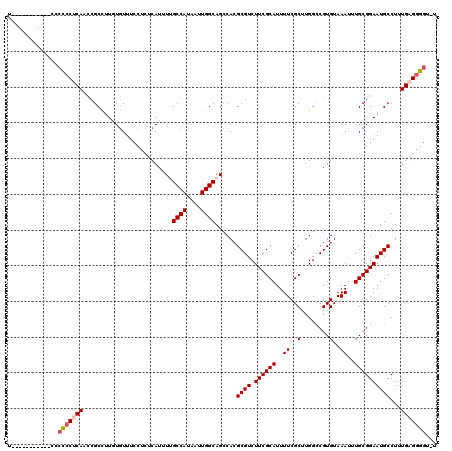
| Location | 10,717,824 – 10,717,932 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.99 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -28.79 |
| Energy contribution | -29.54 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864838 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717824 108 - 23771897 A-ACCCCUCAAAGGCAUUCCGCAAAUUUACACGGCCAAGCGAAAAUGCGAAGACGCGUGGCUGCCAAUUAUGGCCAAAUGAGAGGAAACACAAGGCGGUUGAGGGGGG-----------A .-.((((((((.......((((..........(((((.(((...(((((....)))))...)))......)))))........(....).....))))))))))))..-----------. ( -37.31) >DroSec_CAF1 51826 107 - 1 A-ACCCCUCAAAGGCAUUCCGCAAAUUUACACGGCCAAGCGAAAAUGCGAAGACGCGUGGCUGCCAAUUAUGGCAAGAUGAGAGGAAACACAAGGCGGUUGAGGG-GG-----------A .-.((((((((..((.((((....((((....(((((.(((............))).)))))((((....)))).))))....)))).(....)))..)))))))-).-----------. ( -37.90) >DroSim_CAF1 53689 108 - 1 A-ACCCCUCAAAGGCAUUCCGCAAAUUUACACGGCCAAGCGAAAAUGCGAAGACGCGUGGCUGCCAAUUAUGGCAAGAUGAGAGGAAACACAAGGCGGUUGAGGGGGG-----------A .-.((((((((..((.((((....((((....(((((.(((............))).)))))((((....)))).))))....)))).(....)))..))))))))..-----------. ( -37.90) >DroEre_CAF1 56243 119 - 1 A-ACCCCUCAAAGGCAUUCCGCAAAUUUACACGGCCAAGCGAAAAUGCGAAGACGCGUGGCUGCCAAUUAUGGCAAAAUGAGAGGAAACACAAGGCGGAUGAGGGGGAAUCCGAAGGGGA .-.(((((((.......(((((..........(((((.(((............))).)))))((((....)))).........(....).....))))))))))))...(((....))). ( -42.21) >DroYak_CAF1 65807 117 - 1 A-ACCCCUCAAAGGCAUUCCGCAAAUUUACACGACCAAGCGAAAAUGCGAAGACGCGUGGCUGCCAAUUAUGGCAAAAUGAGAGGAAACAAAAGGCGGAUGGGGGGGGAUCCAAAGGG-- .-.(((((((.......(((((...............(((....(((((....))))).)))((((....)))).........(....).....))))))))))))............-- ( -34.11) >DroAna_CAF1 54094 100 - 1 UGCCUCCUCAGAGGCAUUCCGCAAAUUUACACGGCCAAGCGAAAAUGCGAAGACGCGUGUCUGCCAAUUAUGGCCAAAUGG------AUAAAGAGCCAUUGGGUAG-------------- ((((((....)))))).(((............(((((.(((...(((((....)))))...)))......))))).(((((------........))))))))...-------------- ( -29.90) >consensus A_ACCCCUCAAAGGCAUUCCGCAAAUUUACACGGCCAAGCGAAAAUGCGAAGACGCGUGGCUGCCAAUUAUGGCAAAAUGAGAGGAAACACAAGGCGGUUGAGGGGGG___________A ...((((((((.......((((..........(((((.(((............))).)))))((((....))))....................)))))))))))).............. (-28.79 = -29.54 + 0.75)

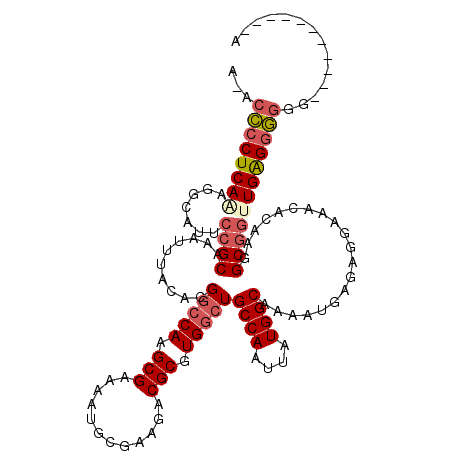
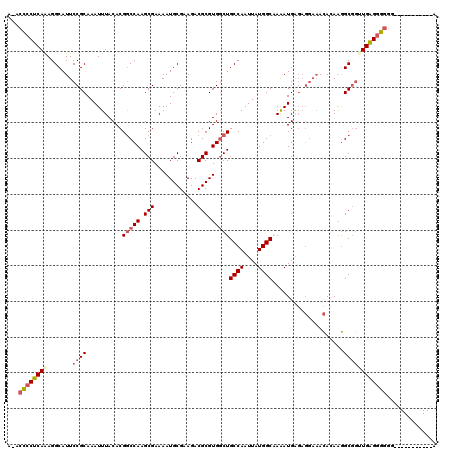
| Location | 10,717,932 – 10,718,050 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.64 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -29.93 |
| Energy contribution | -30.13 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717932 118 + 23771897 UCGCAAAAGAUGGGGGAAA-UCUGGGGAGGUUUCUUUUCAA-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUUUCU ..(.((((((..((((((.-..((.(..(((..(((.....-)))..)))...(((((((((...(((.((((....)))).)))...))))))))).).))..))))))..))))))). ( -40.90) >DroSec_CAF1 51933 115 + 1 UCGCAA-AGAUGGGGGAAA-UCUGAGGAGGUUUCUUUC--A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUAUAU ......-...((..(((((-(((....))))))))..)--)-(((((..(...(((((((((...(((.((((....)))).)))...)))))))))......)..)))))......... ( -37.50) >DroSim_CAF1 53797 115 + 1 UCGCAA-AGAUGGGGGAAA-UCUGGGGAGGUUUCUUUC--A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUAUAU .....(-(((..((((((.-..((.(..(((..((...--.-.))..)))...(((((((((...(((.((((....)))).)))...))))))))).).))..))))))..)))).... ( -38.60) >DroEre_CAF1 56362 115 + 1 UCGCCA-AGAUGGGGGAAA-UCUGGGGAGGUUUCUUUC--A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUUGUU ....((-(((.(((((((.-..((.(..(((..((...--.-.))..)))...(((((((((...(((.((((....)))).)))...))))))))).).))..)))))))..))))).. ( -39.60) >DroYak_CAF1 65924 111 + 1 UCGCAA-AGAUGGGGGGAAUUGUGGGGAGGUUUCUUUU--AAAAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUU------ ......-....(((((((((.(((.(..(((..(((..--..)))..)))...(((((((((...(((.((((....)))).)))...))))))))).).))))))))))))..------ ( -41.50) >DroAna_CAF1 54194 90 + 1 UCA----------GAGAGA-CCAC----AGUCACUU-----------GCUAGAGUUGCCAGGCUUAAUCAAUGUUGACAUUAAUUUGCGUUGGCAAUGCACUUGUUCCUUCC----AUAC ...----------(((.((-(...----.))).)))-----------((....((((((((((..(((.((((....)))).))).)).)))))))))).............----.... ( -18.70) >consensus UCGCAA_AGAUGGGGGAAA_UCUGGGGAGGUUUCUUUC__A_AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUAUAU ...........(((((((..........(((..(((......)))..)))...(((((((((...(((.((((....)))).)))...))))))))).......)))))))......... (-29.93 = -30.13 + 0.20)


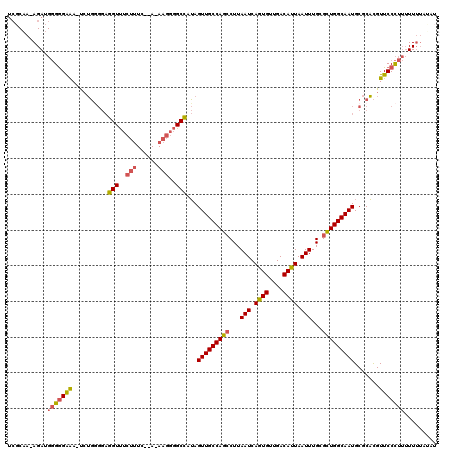
| Location | 10,717,932 – 10,718,050 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.64 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717932 118 - 23771897 AGAAAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-UUGAAAAGAAACCUCCCCAGA-UUUCCCCCAUCUUUUGCGA (((....(((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-)).....((((.((....)).-)))).....)))....... ( -28.20) >DroSec_CAF1 51933 115 - 1 AUAUAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U--GAAAGAAACCUCCUCAGA-UUUCCCCCAUCU-UUGCGA ........((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-)--....((((.((....)).-))))........-...... ( -27.20) >DroSim_CAF1 53797 115 - 1 AUAUAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U--GAAAGAAACCUCCCCAGA-UUUCCCCCAUCU-UUGCGA ........((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-)--....((((.((....)).-))))........-...... ( -27.20) >DroEre_CAF1 56362 115 - 1 AACAAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U--GAAAGAAACCUCCCCAGA-UUUCCCCCAUCU-UGGCGA ........((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-)--..........((((.(((-(.......))))-.)).)) ( -30.00) >DroYak_CAF1 65924 111 - 1 ------AAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUUUU--AAAAGAAACCUCCCCACAAUUCCCCCCAUCU-UUGCGA ------.(((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))))--...............................-...... ( -27.40) >DroAna_CAF1 54194 90 - 1 GUAU----GGAAGGAACAAGUGCAUUGCCAACGCAAAUUAAUGUCAACAUUGAUUAAGCCUGGCAACUCUAGC-----------AAGUGACU----GUGG-UCUCUC----------UGA ....----(((..((.(((((.((((((((..((.((((((((....))))))))..)).)))))........-----------..))))))----.)).-))..))----------).. ( -19.50) >consensus AUAUAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU_U__GAAAGAAACCUCCCCAGA_UUUCCCCCAUCU_UUGCGA .........(((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..))))).......................................... (-19.50 = -20.87 + 1.36)


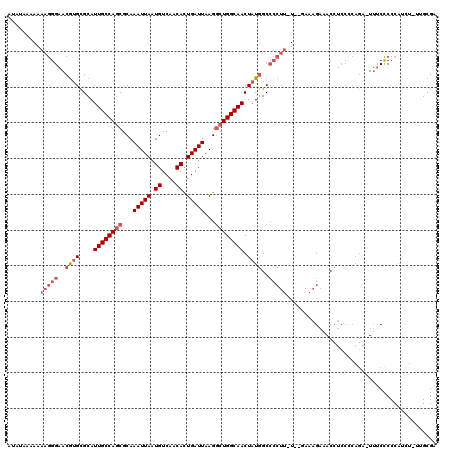
| Location | 10,717,971 – 10,718,073 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.12 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717971 102 + 23771897 A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUUUCUUCAUAUUUUCAGAUUUUU------------GAG-----CA (-(((((..(...(((((((((...(((.((((....)))).)))...)))))))))......)..))))))...............(((((....))------------)))-----.. ( -26.10) >DroSec_CAF1 51969 114 + 1 A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUAUAUAUAUAUUUUCAGAUUUUUGACUCUCAGACUGAG-----CA (-(((((..(...(((((((((...(((.((((....)))).)))...)))))))))......)..))))))...............(((((...((((.....)))))))))-----.. ( -29.70) >DroSim_CAF1 53833 114 + 1 A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUAUAUAUAUAUUUUCAGAUUUUUGACUCUCAGACUGAG-----CA (-(((((..(...(((((((((...(((.((((....)))).)))...)))))))))......)..))))))...............(((((...((((.....)))))))))-----.. ( -29.70) >DroEre_CAF1 56398 108 + 1 A-AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUUGUUGUAUUUUUUCAGAUUUUCGACUCUG-AGCA---------- (-(((((..(...(((((((((...(((.((((....)))).)))...)))))))))......)..))))))...............((((((........))))-))..---------- ( -30.00) >DroYak_CAF1 65961 112 + 1 AAAAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUU-------UAUCUUUUCAGAUUUUUGACUCUU-GACAGAGAGUCUCG (((((((..(...(((((((((...(((.((((....)))).)))...)))))))))......)..))))))).-------.................(((((((-....)))))))... ( -35.30) >DroAna_CAF1 54215 88 + 1 -------GCUAGAGUUGCCAGGCUUAAUCAAUGUUGACAUUAAUUUGCGUUGGCAAUGCACUUGUUCCUUCC----AUAC-----UUAUAAGCUUUUUUGCU----------------CA -------((....((((((((((..(((.((((....)))).))).)).)))))))))).............----....-----.....(((......)))----------------.. ( -17.20) >consensus A_AAGGGGCCAUAGUUGCCAGCCUUAAUCAGUGUUGACAUUAAUUUGCGCUGGCAAUGCGCACGUUCCCUUUUUUUAUAU_UAUAUUUUCAGAUUUUUGACUCU__GAC_GAG_____CA ....(((((....(((((((((...(((.((((....)))).)))...)))))))))......))))).................................................... (-20.73 = -21.12 + 0.39)


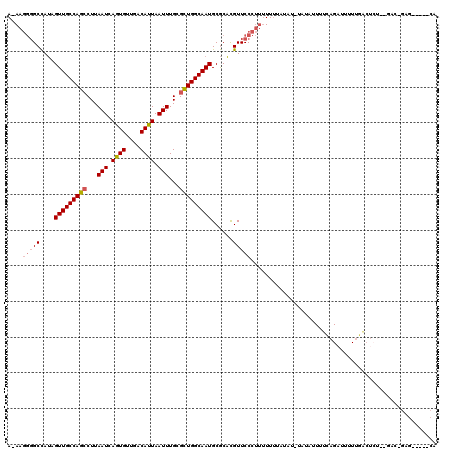
| Location | 10,717,971 – 10,718,073 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.28 |
| Mean single sequence MFE | -28.32 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.87 |
| Covariance contribution | 1.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987488 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10717971 102 - 23771897 UG-----CUC------------AAAAAUCUGAAAAUAUGAAGAAAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U ..-----...------------..........................((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-) ( -25.80) >DroSec_CAF1 51969 114 - 1 UG-----CUCAGUCUGAGAGUCAAAAAUCUGAAAAUAUAUAUAUAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U .(-----(((.......))))...........................((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-) ( -28.20) >DroSim_CAF1 53833 114 - 1 UG-----CUCAGUCUGAGAGUCAAAAAUCUGAAAAUAUAUAUAUAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U .(-----(((.......))))...........................((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-) ( -28.20) >DroEre_CAF1 56398 108 - 1 ----------UGCU-CAGAGUCGAAAAUCUGAAAAAAUACAACAAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU-U ----------...(-((((........)))))................((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..)))))-) ( -30.70) >DroYak_CAF1 65961 112 - 1 CGAGACUCUCUGUC-AAGAGUCAAAAAUCUGAAAAGAUA-------AAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUUUU ...((((((.....-.))))))....((((....)))).-------.(((((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..))))))) ( -38.00) >DroAna_CAF1 54215 88 - 1 UG----------------AGCAAAAAAGCUUAUAA-----GUAU----GGAAGGAACAAGUGCAUUGCCAACGCAAAUUAAUGUCAACAUUGAUUAAGCCUGGCAACUCUAGC------- ((----------------(((......)))))...-----((((----...........)))).((((((..((.((((((((....))))))))..)).)))))).......------- ( -19.00) >consensus UG_____CUC_GUC__AGAGUCAAAAAUCUGAAAAUAUA_AUAUAAAAAAAGGGAACGUGCGCAUUGCCAGCGCAAAUUAAUGUCAACACUGAUUAAGGCUGGCAACUAUGGCCCCUU_U .................................................(((((..((((....((((((((...(((((.((....)).)))))...)))))))).))))..))))).. (-19.50 = -20.87 + 1.36)

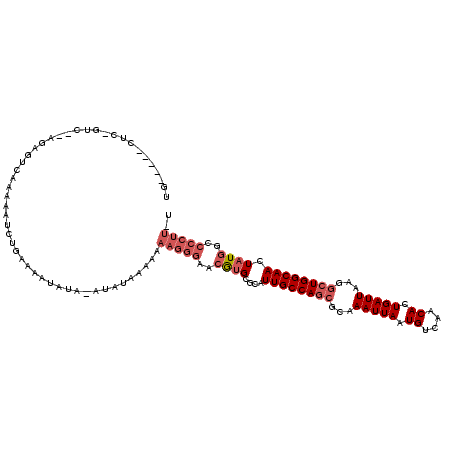
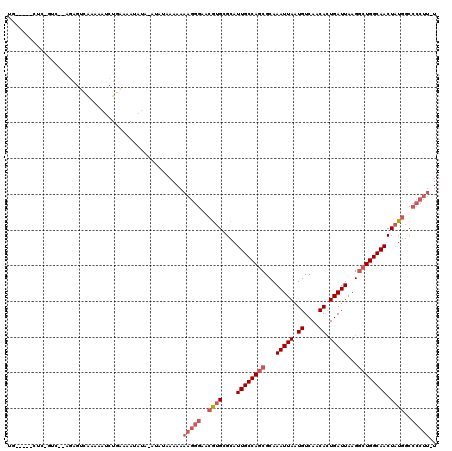
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:31 2006