| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
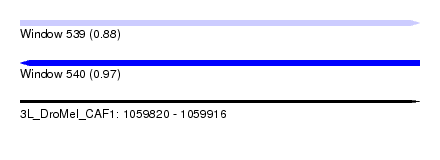
| Location | 1,059,820 – 1,059,916 |
| Length | 96 |
| Max. P | 0.974519 |

| Location | 1,059,820 – 1,059,916 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -26.72 |
| Consensus MFE | -12.62 |
| Energy contribution | -13.68 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
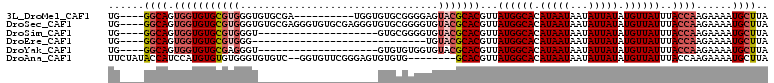
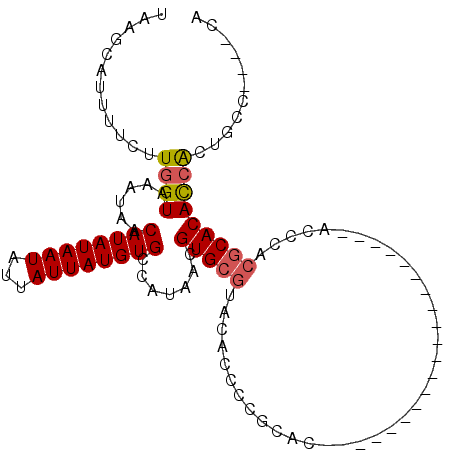
>3L_DroMel_CAF1 1059820 96 + 23771897 UG----GGCAGUGGUGUGCGUGGGUGUGCGA----------UGGUGUGCGGGGAGUACGCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ((----((((.(((((.(((((.((((((.(----------(..((((((.......))))))..)).))))))..........)))))....)))))......)))))) ( -34.00) >DroSec_CAF1 34368 106 + 1 UG----GGCAGUGGUGUGCGUGGGUGUGCGAGGGUGUGCGAGGGUGUGCGGGGUGUACGCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ((----((((.(((((.((((((((((...(..((((((.(..(((((((.......)))))))..).))))))..).))))).)))))....)))))......)))))) ( -33.30) >DroSim_CAF1 36082 86 + 1 UG----GGCAGUGGUGUGCGUGGGU--------------------GUGCGGGGUGUACGCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ((----((((.((((((((...(((--------------------(((((.......))))))))...))))..((((((((...)))))))).))))......)))))) ( -24.40) >DroEre_CAF1 37459 77 + 1 UG----GGCAGUGGUGUGCGUGGG-----------------------------UGUACGCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ((----((((.(((((((((((..-----------------------------..)))))))...((((((.(((((...))))).))))))..))))......)))))) ( -21.30) >DroYak_CAF1 36052 86 + 1 UG----GGCAGUGGUGUGCGAGGGU--------------------GUGUGUGGUGUACGCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ((----((((.((((((((.(..((--------------------(((((((...)))))))))..).))))..((((((((...)))))))).))))......)))))) ( -24.60) >DroAna_CAF1 33570 100 + 1 UUCUAUACCAUCCAUGUGUGUGGGUGUGUC--GGUGUUCGGGAGUGUGUG--------GCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ((((..(((((((((....))))))).)).--((((........((((((--------.((.....)).))))))(((((((...))))))).)))).))))........ ( -22.70) >consensus UG____GGCAGUGGUGUGCGUGGGU____________________GUGCGGGGUGUACGCACGUUAUGGCACAUAAUAAUAUUAUAUGUUAUUUACCAAGAAAAUGCUUA ......((((.(((((((((((.................................)))))))...((((((.(((((...))))).))))))..))))......)))).. (-12.62 = -13.68 + 1.06)
| Location | 1,059,820 – 1,059,916 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Mean single sequence MFE | -15.71 |
| Consensus MFE | -10.83 |
| Energy contribution | -11.06 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
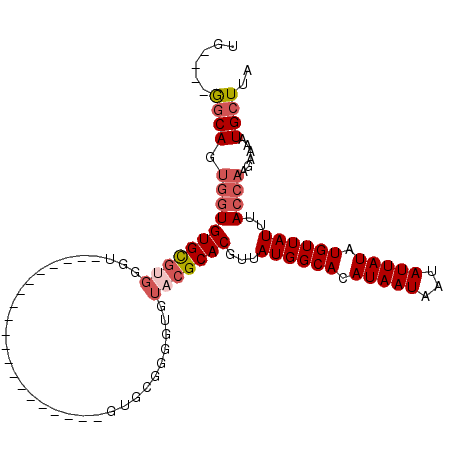
>3L_DroMel_CAF1 1059820 96 - 23771897 UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGCGUACUCCCCGCACACCA----------UCGCACACCCACGCACACCACUGCC----CA ...(((......((((......((((((((...)))))))).......(((((.......)))))))))----------..((........)).......))).----.. ( -18.00) >DroSec_CAF1 34368 106 - 1 UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGCGUACACCCCGCACACCCUCGCACACCCUCGCACACCCACGCACACCACUGCC----CA ...(((......((((.....................(((((......(((((.......)))))......))))).....((........))..)))).))).----.. ( -17.80) >DroSim_CAF1 36082 86 - 1 UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGCGUACACCCCGCAC--------------------ACCCACGCACACCACUGCC----CA ...(((......((((......((((((((...)))))))).......((((((...........--------------------....)))))))))).))).----.. ( -15.16) >DroEre_CAF1 37459 77 - 1 UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGCGUACA-----------------------------CCCACGCACACCACUGCC----CA ...(((......((((......((((((((...)))))))).......((((((...-----------------------------...)))))))))).))).----.. ( -16.70) >DroYak_CAF1 36052 86 - 1 UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGCGUACACCACACAC--------------------ACCCUCGCACACCACUGCC----CA ...(((......((((......((((((((...)))))))).......(((((............--------------------.....))))))))).))).----.. ( -14.03) >DroAna_CAF1 33570 100 - 1 UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGC--------CACACACUCCCGAACACC--GACACACCCACACACAUGGAUGGUAUAGAA ........(((((((((.((((.(........).)))).)))))....((((--------((........((.....)--)......(((......))).)))))))))) ( -12.60) >consensus UAAGCAUUUUCUUGGUAAAUAACAUAUAAUAUUAUUAUGUGCCAUAACGUGCGUACACCCCGCAC____________________ACCCACGCACACCACUGCC____CA ............((((......((((((((...)))))))).......(((((.....................................)))))))))........... (-10.83 = -11.06 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:11 2006