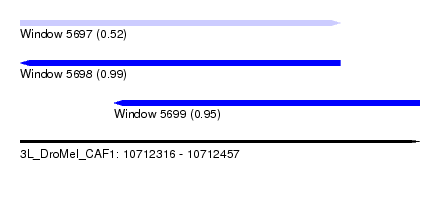
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,712,316 – 10,712,457 |
| Length | 141 |
| Max. P | 0.991822 |

| Location | 10,712,316 – 10,712,429 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -17.79 |
| Energy contribution | -17.57 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.516106 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10712316 113 + 23771897 AAAGCAAGAAAUAAACAAAGGCACAGACAAAAGCAAAGGCAAACAAACACAGCACAAAUAUUAAAGUCAAUUUGCGAAAUGAGAAUGCUGCGAAUGUUUCGCCUUUGGAUUUG ...........................((((..((((((((((((..(.(((((....((((...((......))..))))....))))).)..))))).)))))))..)))) ( -22.20) >DroSec_CAF1 46508 113 + 1 AAAGCAAGAAAUAAACAAAGGCACAGACAAAAGCAAAGGCAAACAAACACAACACAAAUAUUAAAGUCAAUUUGGGAAAUGAGAACGCCGCGAAUGUUUCGCCUUUGGAUUUG ........((((...(((((((..(((((...((...(((..............(((((..........)))))(..........))))))...))))).))))))).)))). ( -19.30) >DroSim_CAF1 48250 113 + 1 AAAGCAAGAAAUAAACAAAGGCACAGACAAAAGCAAAGGCAAACAAACACAACACAAAUAUUAAAGUCAAUUUGCGAAAUGAGAACGCUGCGAAUGUUUCGCCUUUGGAUUUG ........((((...(((((((..(((((...(((..(((.........................))).....(((.........))))))...))))).))))))).)))). ( -19.51) >consensus AAAGCAAGAAAUAAACAAAGGCACAGACAAAAGCAAAGGCAAACAAACACAACACAAAUAUUAAAGUCAAUUUGCGAAAUGAGAACGCUGCGAAUGUUUCGCCUUUGGAUUUG ........((((...(((((((..(((((...((...(((.......((...(.(((((..........))))).)...)).....)))))...))))).))))))).)))). (-17.79 = -17.57 + -0.22)

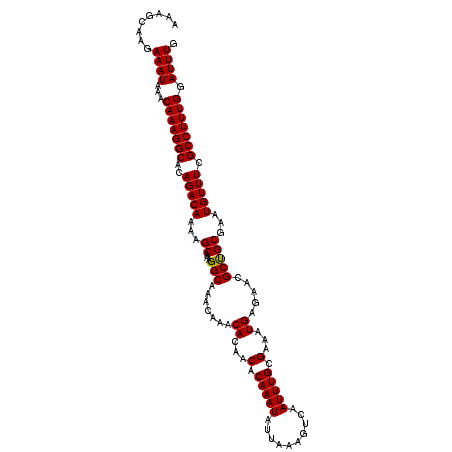

| Location | 10,712,316 – 10,712,429 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 97.64 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -26.53 |
| Energy contribution | -25.87 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10712316 113 - 23771897 CAAAUCCAAAGGCGAAACAUUCGCAGCAUUCUCAUUUCGCAAAUUGACUUUAAUAUUUGUGCUGUGUUUGUUUGCCUUUGCUUUUGUCUGUGCCUUUGUUUAUUUCUUGCUUU ((((..(((((((.(((((..((((((((.............................))))))))..))))))))))))..))))........................... ( -28.95) >DroSec_CAF1 46508 113 - 1 CAAAUCCAAAGGCGAAACAUUCGCGGCGUUCUCAUUUCCCAAAUUGACUUUAAUAUUUGUGUUGUGUUUGUUUGCCUUUGCUUUUGUCUGUGCCUUUGUUUAUUUCUUGCUUU ((((..(((((((.(((((..(((((((...........(((((..........))))))))))))..))))))))))))..))))........................... ( -24.30) >DroSim_CAF1 48250 113 - 1 CAAAUCCAAAGGCGAAACAUUCGCAGCGUUCUCAUUUCGCAAAUUGACUUUAAUAUUUGUGUUGUGUUUGUUUGCCUUUGCUUUUGUCUGUGCCUUUGUUUAUUUCUUGCUUU ((((..(((((((.(((((..(((((((....).....((((((..........))))))))))))..))))))))))))..))))........................... ( -27.20) >consensus CAAAUCCAAAGGCGAAACAUUCGCAGCGUUCUCAUUUCGCAAAUUGACUUUAAUAUUUGUGUUGUGUUUGUUUGCCUUUGCUUUUGUCUGUGCCUUUGUUUAUUUCUUGCUUU ((((..(((((((.(((((..(((((((...........(((((..........))))))))))))..))))))))))))..))))........................... (-26.53 = -25.87 + -0.66)

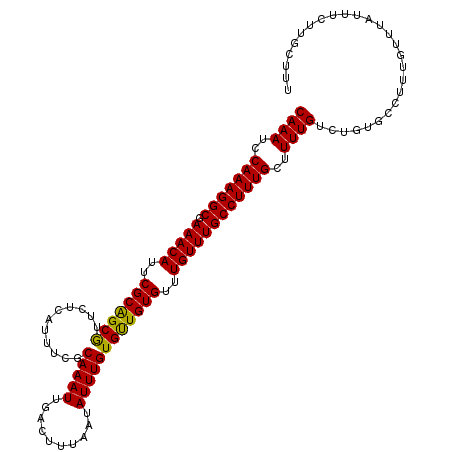

| Location | 10,712,349 – 10,712,457 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -24.73 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10712349 108 - 23771897 GCACACAAUUGCAUAGGCAUUAACAUAUCAAAUCCAAAGGCGAAACAUUCGCAGCAUUCUCAUUUCGCAAAUUGACUUUAAUAUUUGUGCUGUGUUUGUUUGCCUUUG (((......)))......................(((((((.(((((..((((((((.............................))))))))..)))))))))))) ( -27.55) >DroSec_CAF1 46541 104 - 1 ----ACAAUUGCAUAGGCAUUAACAUAUCAAAUCCAAAGGCGAAACAUUCGCGGCGUUCUCAUUUCCCAAAUUGACUUUAAUAUUUGUGUUGUGUUUGUUUGCCUUUG ----.....(((....)))...............(((((((.(((((..(((((((...........(((((..........))))))))))))..)))))))))))) ( -22.50) >DroSim_CAF1 48283 104 - 1 ----ACAAUUGCAUAGGCAUUAACAUAUCAAAUCCAAAGGCGAAACAUUCGCAGCGUUCUCAUUUCGCAAAUUGACUUUAAUAUUUGUGUUGUGUUUGUUUGCCUUUG ----.....(((....)))...............(((((((.(((((..(((((((....).....((((((..........))))))))))))..)))))))))))) ( -25.40) >consensus ____ACAAUUGCAUAGGCAUUAACAUAUCAAAUCCAAAGGCGAAACAUUCGCAGCGUUCUCAUUUCGCAAAUUGACUUUAAUAUUUGUGUUGUGUUUGUUUGCCUUUG .........(((....)))...............(((((((.(((((..(((((((...........(((((..........))))))))))))..)))))))))))) (-24.73 = -24.07 + -0.66)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:20 2006