| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,705,010 – 10,705,100 |
| Length | 90 |
| Max. P | 0.991311 |

| Location | 10,705,010 – 10,705,100 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -22.24 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10705010 90 + 23771897 CUUUUCAUGGGGUUUCAUGGCUGACUUGCGUUCAGCCUCUUUACGCUU-UUCCUCGGCCAUAAGUUGGCUGUUAAAUGCUACUUACGACUG ....((.((((((.....((((((.......)))))).....))((.(-((...((((((.....))))))..))).))..)))).))... ( -23.10) >DroSec_CAF1 39419 90 + 1 CUUUUCAUGGGGUUUCAUGGCUGACUUGCGUUCAGCCUCUUUACGCUU-UUCCUCGGCCAUAAGUUGGCUGUUAAAUGCUACUUACGACUG ....((.((((((.....((((((.......)))))).....))((.(-((...((((((.....))))))..))).))..)))).))... ( -23.10) >DroSim_CAF1 41151 90 + 1 CUUUUCAUGGGGUUUCAUGGCUGACUUGCGUUCAGCCUCUUUACGCUU-UUCCUCGGCCAUAAGUUGGCUGUUAAAUGCUACUUACGACUG ....((.((((((.....((((((.......)))))).....))((.(-((...((((((.....))))))..))).))..)))).))... ( -23.10) >DroEre_CAF1 42755 90 + 1 CUUUUCAUGGGGUUUCAUGGCUGACUGGCGUUCAGCCUCUUUACGCUU-UUCCUCGGCCAUAAGUUGGCUGUUAAAUGCUACUUACGACUG ....((.((((((.....((((((.......)))))).....))((.(-((...((((((.....))))))..))).))..)))).))... ( -23.10) >DroYak_CAF1 52374 90 + 1 CUUUUCAUGGGGUUUCAUGGCUGACUUGC-UUCAGCCUCUUUACGCUUUUUCCUCGGCCAUAAGUUGGCUGUUAAAUGCUACUUACGACUG ....((.((((((.....((((((.....-.)))))).....))((.(((....((((((.....))))))..))).))..)))).))... ( -23.60) >consensus CUUUUCAUGGGGUUUCAUGGCUGACUUGCGUUCAGCCUCUUUACGCUU_UUCCUCGGCCAUAAGUUGGCUGUUAAAUGCUACUUACGACUG ....((.((((((.....((((((.......)))))).....))((........((((((.....))))))......))..)))).))... (-22.24 = -22.24 + 0.00)


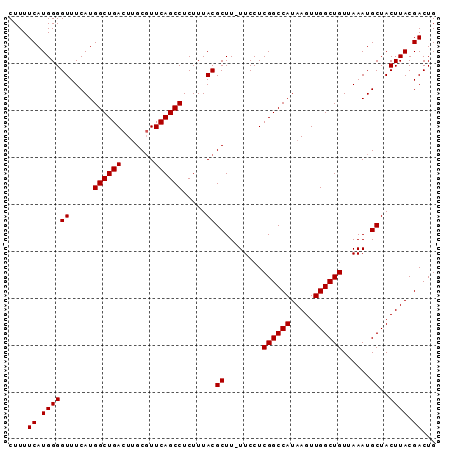
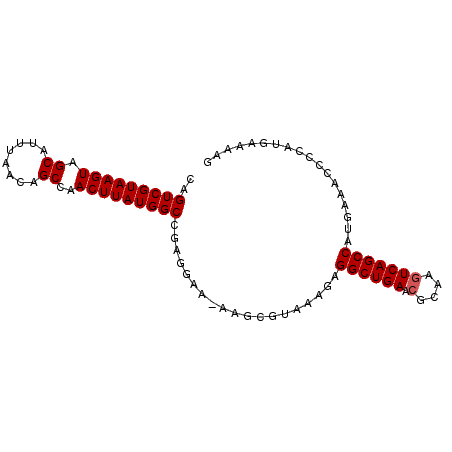
| Location | 10,705,010 – 10,705,100 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 98.67 |
| Mean single sequence MFE | -22.84 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.82 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991311 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10705010 90 - 23771897 CAGUCGUAAGUAGCAUUUAACAGCCAACUUAUGGCCGAGGAA-AAGCGUAAAGAGGCUGAACGCAAGUCAGCCAUGAAACCCCAUGAAAAG ..(((((((((.((........))..)))))))))...((..-...(((.....((((((.(....)))))))))).....))........ ( -24.10) >DroSec_CAF1 39419 90 - 1 CAGUCGUAAGUAGCAUUUAACAGCCAACUUAUGGCCGAGGAA-AAGCGUAAAGAGGCUGAACGCAAGUCAGCCAUGAAACCCCAUGAAAAG ..(((((((((.((........))..)))))))))...((..-...(((.....((((((.(....)))))))))).....))........ ( -24.10) >DroSim_CAF1 41151 90 - 1 CAGUCGUAAGUAGCAUUUAACAGCCAACUUAUGGCCGAGGAA-AAGCGUAAAGAGGCUGAACGCAAGUCAGCCAUGAAACCCCAUGAAAAG ..(((((((((.((........))..)))))))))...((..-...(((.....((((((.(....)))))))))).....))........ ( -24.10) >DroEre_CAF1 42755 90 - 1 CAGUCGUAAGUAGCAUUUAACAGCCAACUUAUGGCCGAGGAA-AAGCGUAAAGAGGCUGAACGCCAGUCAGCCAUGAAACCCCAUGAAAAG ..(((((((((.((........))..)))))))))...((..-...(((.....((((((.(....)))))))))).....))........ ( -21.10) >DroYak_CAF1 52374 90 - 1 CAGUCGUAAGUAGCAUUUAACAGCCAACUUAUGGCCGAGGAAAAAGCGUAAAGAGGCUGAA-GCAAGUCAGCCAUGAAACCCCAUGAAAAG ..(((((((((.((........))..)))))))))...................((((((.-.....)))))).................. ( -20.80) >consensus CAGUCGUAAGUAGCAUUUAACAGCCAACUUAUGGCCGAGGAA_AAGCGUAAAGAGGCUGAACGCAAGUCAGCCAUGAAACCCCAUGAAAAG ..(((((((((.((........))..)))))))))...................((((((.(....))))))).................. (-21.62 = -21.82 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:13 2006