
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,699,431 – 10,699,574 |
| Length | 143 |
| Max. P | 0.947437 |

| Location | 10,699,431 – 10,699,539 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -22.32 |
| Energy contribution | -23.10 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10699431 108 + 23771897 UUGCUGCUGUUGCUUCUUUUUUUUUUCUUUUUAGCCAGCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGCUAAAGGAU (((((((((......................))).))))))..............(((((.((((......(((((((.....))))))).......)))).))))). ( -28.17) >DroSec_CAF1 33842 104 + 1 UUGCUGCUGUUGCUUCUCUUUUUUUUCUUUUUA----GCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUCGCCUUUCGUUUUGAAAGGCUGCCAAUGGUUAAAGGAU ....(((((......................))----)))......((((((((..((....((.......(((((((.....))))))).))....)).)))))))) ( -21.65) >DroSim_CAF1 35463 107 + 1 UUGCUGCUGUUGCUUCUCUUUU-UUUCUUUUUAGCCAGCAAACGAGAUCCUAUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGUUAAAGGAU (((((((((.............-........))).))))))..............(((((.((((......(((((((.....))))))).......)))).))))). ( -25.82) >consensus UUGCUGCUGUUGCUUCUCUUUUUUUUCUUUUUAGCCAGCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGUUAAAGGAU (((((((((......................))).))))))..............(((((.((((......(((((((.....))))))).......)))).))))). (-22.32 = -23.10 + 0.78)

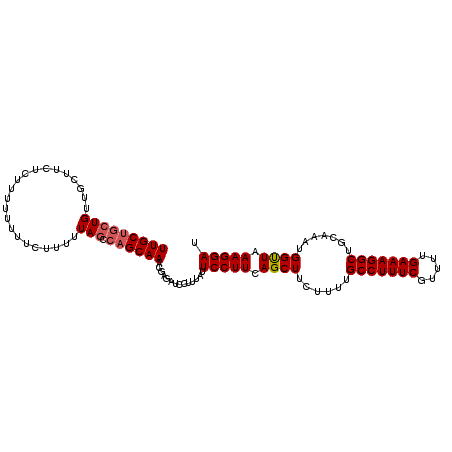
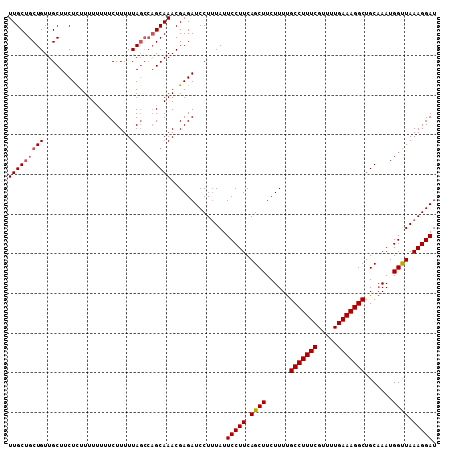
| Location | 10,699,431 – 10,699,539 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -27.17 |
| Consensus MFE | -21.30 |
| Energy contribution | -22.30 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.00 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10699431 108 - 23771897 AUCCUUUAGCCAUUUGCAGCCUUUCAAAACGAAAGGCAAAAGAAGCUGAAGGAAUAAAGGAUCUCGUUUGCUGGCUAAAAAGAAAAAAAAAAGAAGCAACAGCAGCAA .(((((((((..(((...(((((((.....)))))))..)))..)))))))))............(((.(((((((..................)))..))))))).. ( -30.77) >DroSec_CAF1 33842 104 - 1 AUCCUUUAACCAUUGGCAGCCUUUCAAAACGAAAGGCGAAAGAAGCUGAAGGAAUAAAGGAUCUCGUUUGC----UAAAAAGAAAAAAAAGAGAAGCAACAGCAGCAA .(((((((.((.(..((.(((((((.....))))))).......))..).))..)))))))((((.(((.(----(....))....))).)))).((....))..... ( -26.50) >DroSim_CAF1 35463 107 - 1 AUCCUUUAACCAUUUGCAGCCUUUCAAAACGAAAGGCAAAAGAAGCUGAAGGAAUAUAGGAUCUCGUUUGCUGGCUAAAAAGAAA-AAAAGAGAAGCAACAGCAGCAA .(((((((.(..(((...(((((((.....)))))))..)))..).)))))))............(((.(((((((.........-........)))..))))))).. ( -24.23) >consensus AUCCUUUAACCAUUUGCAGCCUUUCAAAACGAAAGGCAAAAGAAGCUGAAGGAAUAAAGGAUCUCGUUUGCUGGCUAAAAAGAAAAAAAAGAGAAGCAACAGCAGCAA ((((((((.((.(..((.(((((((.....))))))).......))..).))..))))))))...(((.((((..........................))))))).. (-21.30 = -22.30 + 1.00)

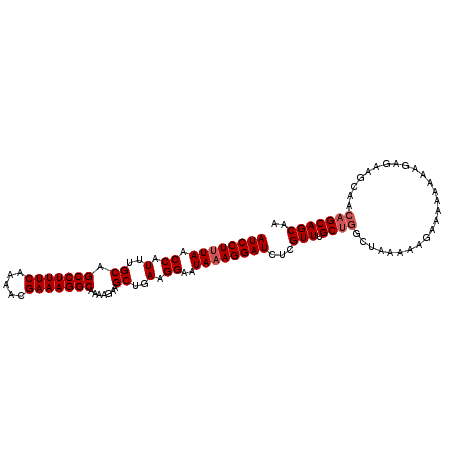
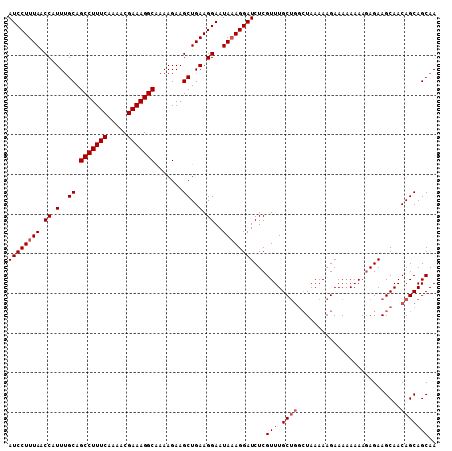
| Location | 10,699,465 – 10,699,574 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 91.98 |
| Mean single sequence MFE | -32.98 |
| Consensus MFE | -28.40 |
| Energy contribution | -28.56 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.803800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10699465 109 + 23771897 CCAGCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGCUAAAGGAUG-----GAGCGGGGAUAGUUGAAGGUGGCAUUGUAGGUAU (((.(.(((...((((((...(((((.((((......(((((((.....))))))).......)))).)))))..-----....)))))).)))...).)))............ ( -31.72) >DroSec_CAF1 33875 106 + 1 ---GCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUCGCCUUUCGUUUUGAAAGGCUGCCAAUGGUUAAAGGAUG-----GAGAGGGGAUAGUUGAAGGUGGCAUUGUAGGUAU ---((..((((...((......((((((((((((((((((((((.....)))))))..(((............))-----).))))))..))))))))).))..))))..)).. ( -31.60) >DroSim_CAF1 35496 109 + 1 CCAGCAAACGAGAUCCUAUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGUUAAAGGAUG-----GAGAGGGGAUAGUUGAAGGUGGCAUUGUAGGUAU ..............((((((..((((((((((((((((((((((.....)))))))......(.(((....))).-----)..)))))).)))))))))......))))))... ( -29.80) >DroEre_CAF1 37422 114 + 1 CCAGCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGUAACAGGGGAGCGGUGAGAGGGGAUAGUUGAAGGUGGCAUUGUGGGUAU ...((..(((((..(((((((((((((.((((((((.(((((((.....)))))))(((.....)))..)))))))).....))))))))....)))))...).))))..)).. ( -34.30) >DroYak_CAF1 46915 109 + 1 CCAGCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUCGAAAGGCUGCAAAUGGUUGCAGGAUG-----GAGGGGGGAUAGUUGAAGGUGGCAUUGUAGGUAU ...((..((((...((......(((((((((...(((.(((((((((((....))((((((....))))))))))-----))))).))).))))))))).))..))))..)).. ( -37.50) >consensus CCAGCAAACGAGAUCCUUUAUUCCUUCAGCUUCUUUUGCCUUUCGUUUUGAAAGGCUGCAAAUGGUUAAAGGAUG_____GAGAGGGGAUAGUUGAAGGUGGCAUUGUAGGUAU ...((..((((...((......((((((((((((((((((((((.....))))))).((.....)).................)))))).))))))))).))..))))..)).. (-28.40 = -28.56 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:08 2006