
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,696,607 – 10,696,727 |
| Length | 120 |
| Max. P | 0.737484 |

| Location | 10,696,607 – 10,696,727 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -30.71 |
| Energy contribution | -31.83 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
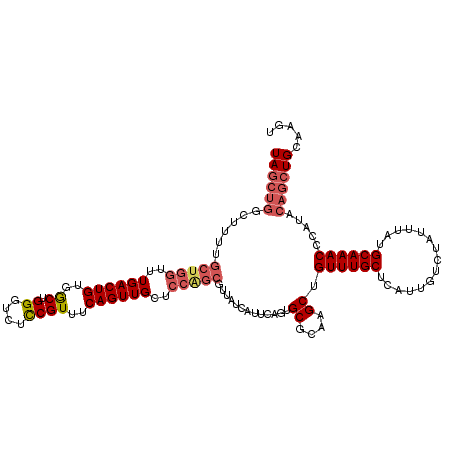
>3L_DroMel_CAF1 10696607 120 + 23771897 UAGCUGGCUUUUGCUGGUUUGACUGUGGCUGGAUCUCCGUUUCAGUUGCUCCAGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACCCAUACAGCUGCAAGU ((((((......(((((..((((((..((.((....))))..))))))..))))).............((....)).((((((...............)))))).....))))))..... ( -37.06) >DroSec_CAF1 31014 120 + 1 UAGCUGGCUUUUGCUGGUUUGACUGUGGCUGGGUCUCCGUUUCAGUUGCUCCAGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACCCAUACAGCUGCAAGU ((((((......(((((..((((((..((.((....))))..))))))..))))).............((....)).((((((...............)))))).....))))))..... ( -36.16) >DroSim_CAF1 32622 120 + 1 UAGCUGGCUUUUGCUGGUUUGACUGUGGCUGGGUCUCCGUUUCAGUUGCUCCAGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACCCAUACAGCUGCAAGU ((((((......(((((..((((((..((.((....))))..))))))..))))).............((....)).((((((...............)))))).....))))))..... ( -36.16) >DroEre_CAF1 34546 120 + 1 UAGCUGGCUUUUGCUGGUUUGACUGUGGCUGGGUCUCCGUUUCAGUUGCUCCAGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACCCAUACAGCUGCAAGU ((((((......(((((..((((((..((.((....))))..))))))..))))).............((....)).((((((...............)))))).....))))))..... ( -36.16) >DroYak_CAF1 43975 120 + 1 UAGCUGGCUUUUGCUGGCUUGACUGUGGCUGGGACUCCGUUUCAGUUGCUCCGGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACCCAUACAGCUGCAAGU .(((..((....))..)))....((..((((((((..(......)..).))).((((.......(((((.(((((...))))).))))).......)))).........))))..))... ( -36.24) >DroAna_CAF1 31264 105 + 1 UAG---------------UUGACUGGGACUGGGUCCUCGUUUCAGUUGCUCCAGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACACAUACAGAUGCAGAA ...---------------..(((((..((.((....))))..)))))......(((((..........((....))(((((((...............)))))))......))))).... ( -25.66) >consensus UAGCUGGCUUUUGCUGGUUUGACUGUGGCUGGGUCUCCGUUUCAGUUGCUCCAGCGUUAUCAUUCAGUGCGCAAGCUGUUUGCUCAUUGUCUAUUUAUGCAAACCCAUACAGCUGCAAGU ((((((......(((((..((((((..((.((....))))..))))))..))))).............((....)).((((((...............)))))).....))))))..... (-30.71 = -31.83 + 1.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:03:02 2006