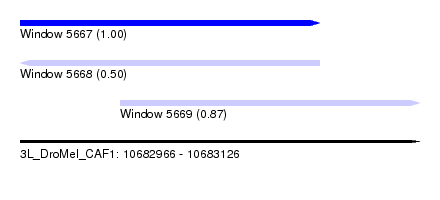
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,682,966 – 10,683,126 |
| Length | 160 |
| Max. P | 0.997801 |

| Location | 10,682,966 – 10,683,086 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.01 |
| Mean single sequence MFE | -45.20 |
| Consensus MFE | -40.78 |
| Energy contribution | -40.65 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10682966 120 + 23771897 CUACGGCGGUGGCGCGUCGUCGUUGUGUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCC ...(((((((((((.((....(((((((...))))))).........((((((....((....))....))))))))))))))))))).((((((......))))))............. ( -42.40) >DroSec_CAF1 17297 117 + 1 CUGCGGCGGUGGCGCGU---CGUUGUGUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGACUCCAAGUUGGAGCCUCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCC .((((((((((((((((---....((((.....((.........)).....))))..))))).((((.....))))...)))))))))))(((((......))))).............. ( -42.00) >DroSim_CAF1 18538 117 + 1 CUGCGGCGGUGGCGCGU---CGUUGUGUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAUCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCC .(((((((((((((.((---((((((((...)))))))...........((((....((....))....))))))).)))))))))))))(((((......))))).............. ( -47.40) >DroEre_CAF1 17932 117 + 1 UUGCGGCGGUGGCGCGU---CGUUGUGUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCGCUUAAGCUUGUCC .(((((((((((((.((---.(((((((...))))))).........((((((....((....))....)))))))))))))))))))))(((((......)))))((....))...... ( -48.40) >DroYak_CAF1 29341 117 + 1 UUGCGGCGGUGGCGCGU---CGUUGUGUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUGAGCUUGUCC .(((((((((((((.((---.(((((((...))))))).........((((((....((....))....)))))))))))))))))))))(((((......))))).............. ( -46.90) >DroAna_CAF1 17698 116 + 1 -AGCGGUGGCAGCGCGU---CGUUGUAUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCUGCUGUCGCUGCAGCUGCUCGUGUGCAGCUCUUAAGCCUGUCC -.(((((((((((((..---.(((((((...))))))).........((((((....((....))....)))))))).)))))))))))((((((......))))))............. ( -44.10) >consensus CUGCGGCGGUGGCGCGU___CGUUGUGUGUUAUACAAUAAUUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCC .(((((((((((((.((....(((((((...))))))).........((((((....)).))))(((.....))))))))))))))))))(((((......))))).............. (-40.78 = -40.65 + -0.13)


| Location | 10,682,966 – 10,683,086 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.01 |
| Mean single sequence MFE | -36.15 |
| Consensus MFE | -30.74 |
| Energy contribution | -31.02 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10682966 120 - 23771897 GGACAAGCUUAAGAGCUGCAAACGAGCAGCUGCAGCGGUAGCGGCUCCAACUUGGACUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACACACAACGACGACGCGCCACCGCCGUAG .............((((((......))))))((.(((((.(((..(((.....)))....))).((((((((....))).(..((((.......))))..)....)))))))))).)).. ( -33.70) >DroSec_CAF1 17297 117 - 1 GGACAAGCUUAAGAGCUGCAAACGAGCAGCUGCAGCGGUAGAGGCUCCAACUUGGAGUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACACACAACG---ACGCGCCACCGCCGCAG .............((((((......))))))((.(((((...((((((.....)))))).....((((((((....))).(..((((.......))))..---).)))))))))).)).. ( -38.80) >DroSim_CAF1 18538 117 - 1 GGACAAGCUUAAGAGCUGCAAACGAGCAGCUGCAGCGGUAGCGGAUCCAACUUGGACUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACACACAACG---ACGCGCCACCGCCGCAG .............((((((......))))))((.(((((.((((((((.....))).)).))).((((((((....))).(..((((.......))))..---).)))))))))).)).. ( -37.70) >DroEre_CAF1 17932 117 - 1 GGACAAGCUUAAGCGCUGCAAACGAGCAGCUGCAGCGGUAGCGGCUCCAACUUGGACUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACACACAACG---ACGCGCCACCGCCGCAA ......((....))(((((......)))))(((.(((((.(((..(((.....)))....))).((((((((....))).(..((((.......))))..---).)))))))))).))). ( -36.90) >DroYak_CAF1 29341 117 - 1 GGACAAGCUCAAGAGCUGCAAACGAGCAGCUGCAGCGGUAGCGGCUCCAACUUGGACUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACACACAACG---ACGCGCCACCGCCGCAA .............((((((......))))))((.(((((.(((..(((.....)))....))).((((((((....))).(..((((.......))))..---).)))))))))).)).. ( -35.90) >DroAna_CAF1 17698 116 - 1 GGACAGGCUUAAGAGCUGCACACGAGCAGCUGCAGCGACAGCAGCUCCAACUUGGACUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACAUACAACG---ACGCGCUGCCACCGCU- .....(((.....((((((......))))))((((((.(((((.((((.....)))........).))))).........(..((((((...))))))..---)..))))))....)))- ( -33.90) >consensus GGACAAGCUUAAGAGCUGCAAACGAGCAGCUGCAGCGGUAGCGGCUCCAACUUGGACUCACGCAGGUGCUGAAAACUUAAUUAUUGUAUAACACACAACG___ACGCGCCACCGCCGCAG .............((((((......))))))((.(((((.(((..(((.....)))....))).((((((((....)))....((((.......)))).......)))))))))).)).. (-30.74 = -31.02 + 0.28)
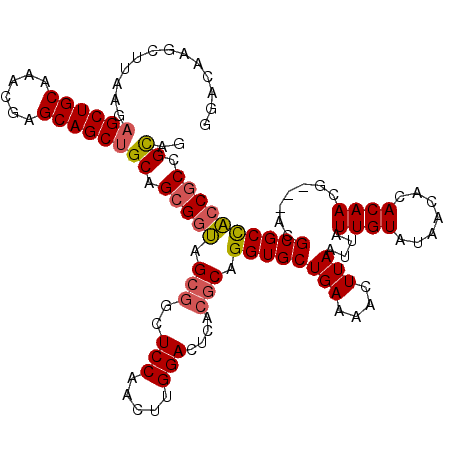
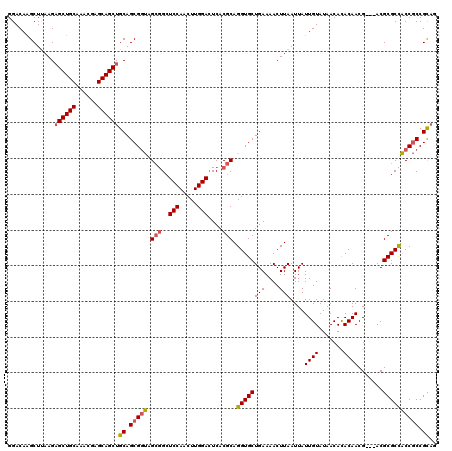
| Location | 10,683,006 – 10,683,126 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.69 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -36.62 |
| Energy contribution | -36.65 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10683006 120 + 23771897 UUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCCAGACAACCAGCCCGAUGAGCCAAUGAGCGGCUCCAAAGAU ....(..((((((....)).))))..)...(((((((((((...(((((((........)))))))......(((..((..(........)..))..))).....))))))))))).... ( -42.50) >DroSec_CAF1 17334 118 + 1 UUAAGUUUUCAGCACCUGCGUGACUCCAAGUUGGAGCCUCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCCAGACAACCAGCCCGAU--GCCAAUGAGCGGCUCCAAAGAU ...((((....((....))..)))).....((((((((......((....)).(((((((.((((((....)))((((....)))).........)--)).))))))))))))))).... ( -35.20) >DroSim_CAF1 18575 118 + 1 UUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAUCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCCAGACAACCAGCCCGAU--GCCAAUGAGCGGCUCCAAAGAU .........((((....(((.((.(((.....))))))))....)))).(((((((((((.((((((....)))((((....)))).........)--)).)))))))))))........ ( -36.70) >DroEre_CAF1 17969 118 + 1 UUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCGCUUAAGCUUGUCCAGACAACCAGCCCGAU--GCCAAUGAGCGGCUCCAAAGAC ....(..((((((....)).))))..)...(((((((((((......((((((((......)))))(((...(.((((....)))).))))....)--)).....))))))))))).... ( -41.90) >DroYak_CAF1 29378 118 + 1 UUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUGAGCUUGUCCAGACAACCAGCCCGAU--GCCAAUGAGCGGCUCCAAAGAU ....(..((((((....)).))))..)...(((((((((((......((((((((......)))))..(((.((((((....)))...))).))))--)).....))))))))))).... ( -42.60) >DroAna_CAF1 17734 118 + 1 UUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCUGCUGUCGCUGCAGCUGCUCGUGUGCAGCUCUUAAGCCUGUCCAGACAGCGAGCCACAA--AAGCAGGAGCGGUUCGGGACUC ...((((((..((.(((((.((..(((.....)))((((((((((((..((((((......))))))....))).......)))))).)))..)).--..))))).)).....)))))). ( -41.71) >consensus UUAAGUUUUCAGCACCUGCGUGAGUCCAAGUUGGAGCCGCUACCGCUGCAGCUGCUCGUUUGCAGCUCUUAAGCUUGUCCAGACAACCAGCCCGAU__GCCAAUGAGCGGCUCCAAAGAU ....(..((((((....)).))))..)...(((((((((((...(((..((((((......))))))....)))((((....))))...................))))))))))).... (-36.62 = -36.65 + 0.03)

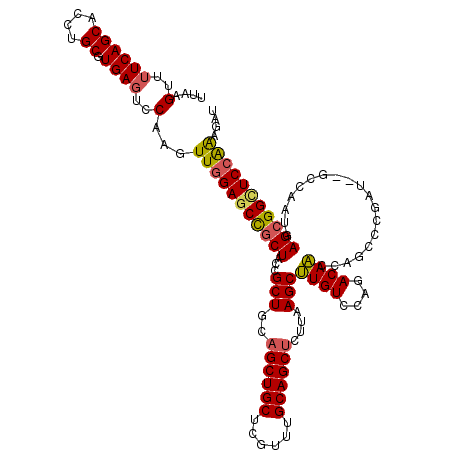
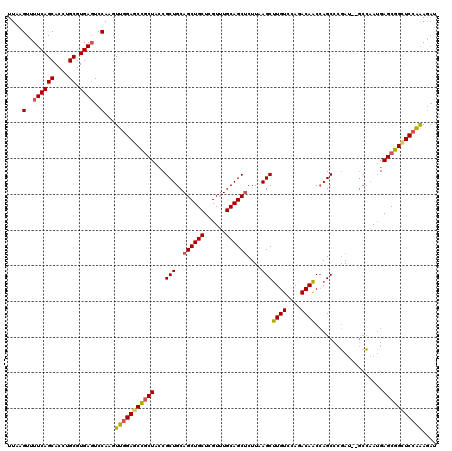
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:50 2006