
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,682,580 – 10,682,735 |
| Length | 155 |
| Max. P | 0.848181 |

| Location | 10,682,580 – 10,682,695 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.73 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10682580 115 + 23771897 UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCUCUUUUUCUCCACCUGUUCUCCCAGUUACUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGCUACUAAA-----AAACUGGUUACCC .((((((((.((((........((((.((...............)))))).......((.(((......)))...))..))))...))))))))..((((..-----....))))..... ( -23.16) >DroSec_CAF1 16922 115 + 1 UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCUCUUUUUCUCCACCUGUUCUCCCAGUUACUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGCUACUAAA-----AAACUGGUUUUCC .((((((((.((((........((((.((...............)))))).......((.(((......)))...))..))))...))))))))..((((..-----....))))..... ( -23.16) >DroSim_CAF1 18152 115 + 1 UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCUCUUUUUCUCCACCUGUUCUCCCAGUUACUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGCUACUAAA-----AAACUGGUUUCCC .((((((((.((((........((((.((...............)))))).......((.(((......)))...))..))))...))))))))..((((..-----....))))..... ( -23.16) >DroEre_CAF1 17547 115 + 1 UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCUCUUUUGCUCCACCUGUUCUCCCAGUUACUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGCUACUAAA-----AAACUGGUUUUCC .((((((((.((((........((((.((...(......)....)))))).......((.(((......)))...))..))))...))))))))..((((..-----....))))..... ( -23.20) >DroYak_CAF1 28946 120 + 1 UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCUCUUUUUCUCCACCUGUUCUCCCAGUUACUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGCUACUAAAAAAAAAAACUGGUUUUCC .((((((((.((((........((((.((...............)))))).......((.(((......)))...))..))))...)))))))).......................... ( -22.96) >DroAna_CAF1 17306 96 + 1 UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCCCC---------------CCCUCGAUGGCUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGGGCCUUCG-----AAACAGGUU---- .((((((((.((((((((((.......(((.((...---------------.....)).)))..))))))(.....)..))))...))))))))(((((...-----....)))))---- ( -25.50) >consensus UUAGGCGCCCGUGCUAAUUGUCACAGUGUCUUCUCUUUUUCUCCACCUGUUCUCCCAGUUACUUCAAUUAGUUUUACAUGCACUUUGGCGCCUGCUACUAAA_____AAACUGGUUUUCC .((((((((.((((((((((....((((...............................)))).))))))(.....)..))))...)))))))).......................... (-20.70 = -20.73 + 0.03)


| Location | 10,682,580 – 10,682,695 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -24.18 |
| Energy contribution | -25.18 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660412 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10682580 115 - 23771897 GGGUAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAGAAAAAGAGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ((....)).....-----.........(((((((...((((..(.....)((((((......(((......)))(((...............))).....)))))))))).))))))).. ( -29.86) >DroSec_CAF1 16922 115 - 1 GGAAAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAGAAAAAGAGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ((((((....)))-----)))......(((((((...((((..(.....)((((((......(((......)))(((...............))).....)))))))))).))))))).. ( -28.66) >DroSim_CAF1 18152 115 - 1 GGGAAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAGAAAAAGAGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ((....)).....-----.........(((((((...((((..(.....)((((((......(((......)))(((...............))).....)))))))))).))))))).. ( -32.86) >DroEre_CAF1 17547 115 - 1 GGAAAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAGCAAAAGAGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ((((((....)))-----)))......(((((((...((((..(.....)((((((......(((......)))(((...(......)....))).....)))))))))).))))))).. ( -28.80) >DroYak_CAF1 28946 120 - 1 GGAAAACCAGUUUUUUUUUUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAGAAAAAGAGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ......((((((.((((..(((((.(((.((((....)))).)))...)))))..)))).))))))......(((((........(......).(((((........))))).))))).. ( -30.90) >DroAna_CAF1 17306 96 - 1 ----AACCUGUUU-----CGAAGGCCCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGCCAUCGAGGG---------------GGGGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ----..(((....-----...)))...(((((((...((((........((..((((.....))))..)---------------)((.......))..........)))).))))))).. ( -27.10) >consensus GGAAAACCAGUUU_____UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAGAAAAAGAGAAGACACUGUGACAAUUAGCACGGGCGCCUAA ...........................(((((((...((((..(.....)((((((......(((......)))(((...............))).....)))))))))).))))))).. (-24.18 = -25.18 + 1.00)

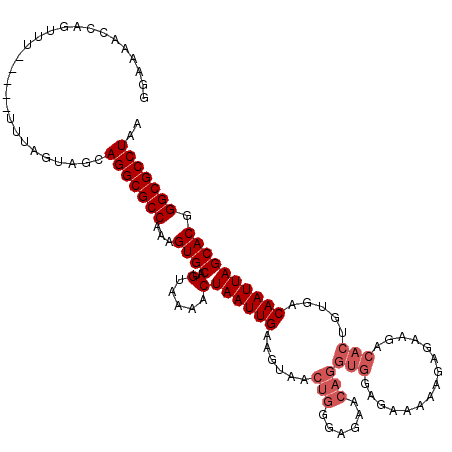
| Location | 10,682,620 – 10,682,735 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.44 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848181 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10682620 115 - 23771897 CUCAUUUCUUCGACAAGAAAGAAAGAGAGAAGCGGCGAUUGGGUAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAG (((.(((((((.....).))))))))).....(.((..(((..(..((((((.-----.(((((.(((.((((....)))).)))...))))).......))))))..)..))))).).. ( -25.80) >DroSec_CAF1 16962 115 - 1 CUCAUUUCUCCGACAAGAUAGAAAGAGAGAGAAGGCGAUUGGAAAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAG (((.(((((((.............).))))))...((.(((.....((((((.-----.(((((.(((.((((....)))).)))...))))).......)))))).....))).))))) ( -23.82) >DroSim_CAF1 18192 115 - 1 CUCAUUUCUCCAACAAGAAAGAAAGAGAGAGAAGGCGAUUGGGAAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAG .((((((((((....((...................((((((....)))))).-----.(((((.(((.((((....)))).)))...))))).........)).))))))...)))).. ( -26.90) >DroEre_CAF1 17587 113 - 1 CUCAUUUCUCCGUCAAGAAAAA--GAGAGAGAUGGCGAUUGGAAAACCAGUUU-----UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAG .((((((((((...........--).)))))))))((.(((.....((((((.-----.(((((.(((.((((....)))).)))...))))).......)))))).....))).))... ( -26.60) >DroYak_CAF1 28986 118 - 1 CUCAUUUCUCGAUCAAGAAAGA--GAGAGAGCAAACGUUUGGAAAACCAGUUUUUUUUUUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAG (((..(((((..((......))--..)))))....((..((.....((((((.((((..(((((.(((.((((....)))).)))...)))))..)))).)))))).....))..))))) ( -33.60) >consensus CUCAUUUCUCCGACAAGAAAGAAAGAGAGAGAAGGCGAUUGGAAAACCAGUUU_____UUUAGUAGCAGGCGCCAAAGUGCAUGUAAAACUAAUUGAAGUAACUGGGAGAACAGGUGGAG (((.((((((..............)))))).....((.(((.....((((((.......(((((.(((.((((....)))).)))...))))).......)))))).....))).))))) (-24.88 = -24.72 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:47 2006