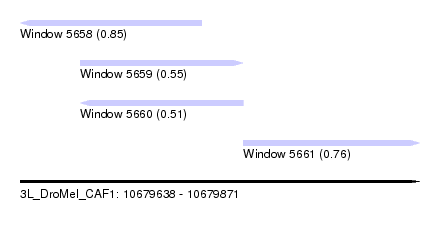
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,679,638 – 10,679,871 |
| Length | 233 |
| Max. P | 0.849267 |

| Location | 10,679,638 – 10,679,744 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 95.47 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849267 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
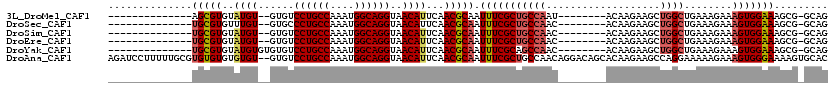
>3L_DroMel_CAF1 10679638 106 - 23771897 UGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAAUACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCGGCAGCAACAUUCGUAGAAUGGGUUAGAUCCGUCUGCAAC ..(((((...(((((.(((.(((...((((((....((......))(((..............)))))))))...)))))).)))))((......)).)))))... ( -29.74) >DroSec_CAF1 14075 106 - 1 UGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAACACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCGGCAGCAACAGCCGUAGAAUGGGAUAGAUCCGUCUGCAGC ..(((((...(((((..(((..((((.(.((((.(.......).)))).)...)))))))....(((((.......))))).)))))(((....))).)))))... ( -30.90) >DroSim_CAF1 14723 106 - 1 UGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAACACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCGGCAGCAACAGCCGUAGAAUGGGAUAGAUCCGUCUGCAGC ..(((((...(((((..(((..((((.(.((((.(.......).)))).)...)))))))....(((((.......))))).)))))(((....))).)))))... ( -30.90) >DroEre_CAF1 14650 106 - 1 UGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAACACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCGGCAGCUACAUCCGUAGAAUGGGAAAGAUCCGUCUGCAGC ..(((((...(((((.(((.......((((((..(....)..(((..(((........)))..)))))))))......))).)))))(((....))).)))))... ( -30.52) >DroYak_CAF1 26022 106 - 1 UGGCAGGUAACAUUCAACGCAAUUUCGCAGCCAACACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCGGCAGCAACAUCCGUAGAAUGGUAAGGAUCCGUCUGCGGC ..(((((...(((((.(((((.((((.((((((.(.......).))))))...)))).))....((....))......))).)))))....(....).)))))... ( -28.60) >consensus UGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAACACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCGGCAGCAACAUCCGUAGAAUGGGAUAGAUCCGUCUGCAGC ..(((((...(((((.(((.......((((((..(....)..(((..(((........)))..)))))))))......))).)))))((......)).)))))... (-27.44 = -27.68 + 0.24)

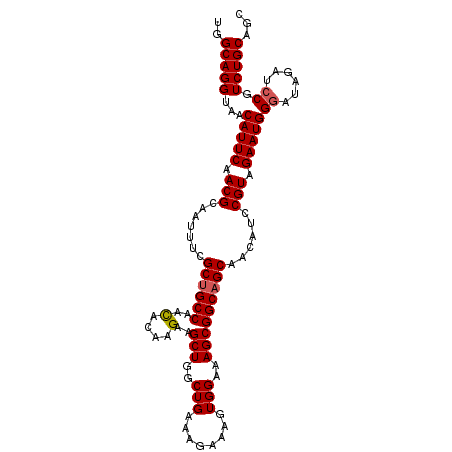
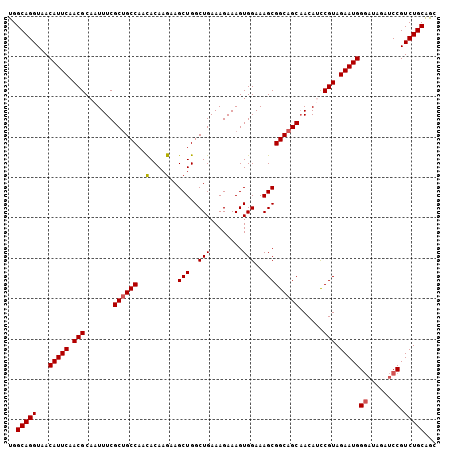
| Location | 10,679,673 – 10,679,768 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.84 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.78 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10679673 95 + 23771897 CUGC-CGCUUUCCACUUUCUUUCAGCCAGCUUCUUGU--------AUUGGCAGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGACAC--ACAUACACGCU-------------- ((((-((((..............)))..((.....))--------...))))).......((((.(.((((..((((((....))))))....--)))).))))).-------------- ( -26.14) >DroSec_CAF1 14110 95 + 1 CUGC-CGCUUUCCACUUUCUUUCAGCCAGCUUCUUGU--------GUUGGCAGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGGCAC--ACAAACACGCA-------------- ....-((((...............((((((.......--------))))))))))....(((((....(((..((((((....))))))..))--).....)))))-------------- ( -30.56) >DroSim_CAF1 14758 95 + 1 CUGC-CGCUUUCCACUUUCUUUCAGCCAGCUUCUUGU--------GUUGGCAGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGACAC--ACAUACACGCA-------------- ....-((((...............((((((.......--------))))))))))....(((((.(.((((..((((((....))))))....--)))).))))))-------------- ( -30.56) >DroEre_CAF1 14685 95 + 1 CUGC-CGCUUUCCACUUUCUUUCAGCCAGCUUCUUGU--------GUUGGCAGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGACAC--ACAUACACGCA-------------- ....-((((...............((((((.......--------))))))))))....(((((.(.((((..((((((....))))))....--)))).))))))-------------- ( -30.56) >DroYak_CAF1 26057 97 + 1 CUGC-CGCUUUCCACUUUCUUUCAGCCAGCUUCUUGU--------GUUGGCUGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGACACACACAUACACGCA-------------- ....-..........((((...((((((((.......--------)))))))).)))).(((((((..(((..((((((....))))))..)))..))...)))))-------------- ( -31.70) >DroAna_CAF1 14323 118 + 1 GUGCACUUUUCCCACUUUCUUUUUCCUGGCUUCUUGUGCUGUCCUGUUGGCAGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGACAC--ACACACACACACGCAAAAAGGAUCU .......................((((.........(((((.((....)))))))...((((((((..(((..((((((....))))))..))--)..)).....))))))..))))... ( -31.60) >consensus CUGC_CGCUUUCCACUUUCUUUCAGCCAGCUUCUUGU________GUUGGCAGCGAAAUUGCGUUGAAUGUUACCUGCCAUUUGGCAGGACAC__ACAUACACGCA______________ ...................((((.((((((...............))))))...))))..((((.(.((((..((((((....))))))......)))).)))))............... (-22.36 = -22.78 + 0.42)

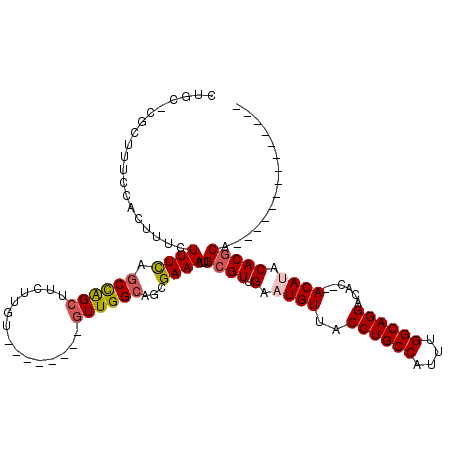
| Location | 10,679,673 – 10,679,768 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.84 |
| Mean single sequence MFE | -33.34 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.88 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.73 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.509559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10679673 95 - 23771897 --------------AGCGUGUAUGU--GUGUCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAAU--------ACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCG-GCAG --------------.((((..((((--....((((((....))))))..))))...)))).......(((((...--------.((......))(((..............))))-)))) ( -31.14) >DroSec_CAF1 14110 95 - 1 --------------UGCGUGUUUGU--GUGCCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAAC--------ACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCG-GCAG --------------(((((....((--((..((((((....))))))..))))...)))))......(((((..(--------....)..(((..(((........)))..))))-)))) ( -33.50) >DroSim_CAF1 14758 95 - 1 --------------UGCGUGUAUGU--GUGUCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAAC--------ACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCG-GCAG --------------(((((..((((--....((((((....))))))..))))...)))))......(((((..(--------....)..(((..(((........)))..))))-)))) ( -32.60) >DroEre_CAF1 14685 95 - 1 --------------UGCGUGUAUGU--GUGUCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAAC--------ACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCG-GCAG --------------(((((..((((--....((((((....))))))..))))...)))))......(((((..(--------....)..(((..(((........)))..))))-)))) ( -32.60) >DroYak_CAF1 26057 97 - 1 --------------UGCGUGUAUGUGUGUGUCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCAGCCAAC--------ACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCG-GCAG --------------(((((....((((....((((((....))))))..))))...))))).((((.((((((.(--------.......).))))))...))))((.(....).-)).. ( -35.20) >DroAna_CAF1 14323 118 - 1 AGAUCCUUUUUGCGUGUGUGUGUGU--GUGUCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAACAGGACAGCACAAGAAGCCAGGAAAAAGAAAGUGGGAAAAGUGCAC ...((((..((((((.((...((((--....((((((....))))))..)))).))))))))....((((((....)).))))..........))))........(((.(.....).))) ( -35.00) >consensus ______________UGCGUGUAUGU__GUGUCCUGCCAAAUGGCAGGUAACAUUCAACGCAAUUUCGCUGCCAAC________ACAAGAAGCUGGCUGAAAGAAAGUGGAAAGCG_GCAG ..............(((((..((((......((((((....))))))..))))...))))).(((((((((((...................))))........)))))))......... (-24.26 = -24.88 + 0.61)
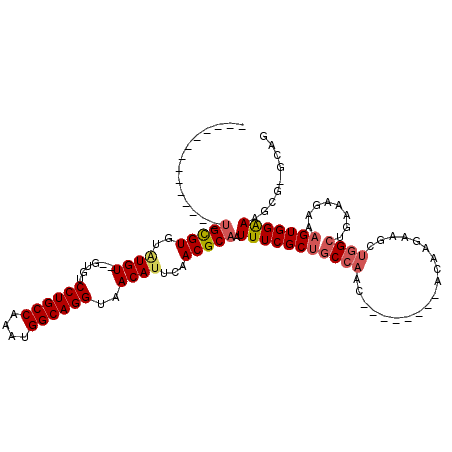
| Location | 10,679,768 – 10,679,871 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 85.60 |
| Mean single sequence MFE | -34.48 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10679768 103 + 23771897 GGGUCCAGGAUCCCUUUUAACCCGGGAUUCUGCACCACCAUC------UUCCUUCCAUCCUCUCCUCUCCACCAGUCGAGUAUUAAGUAAAUGUUUGGGGCCUGUAGGA .(((.(((((((((.........))))))))).)))......------..............(((((.((.((((.((..(((...)))..)).))))))...).)))) ( -26.30) >DroSec_CAF1 14205 100 + 1 GGGUCCAGGAUCCCUUUUAACCCGGGAUUCUGCACCACCAUC------GGCCUCCACUCCUCUCGUCUCCGCCAGUCGAGUAUUAAGUAAAUGUU-GGGGCC--UAGGA .(((.(((((((((.........))))))))).))).((...------((((((.((....((((.((.....)).))))............)).-))))))--..)). ( -35.09) >DroSim_CAF1 14853 100 + 1 GGGUCCAGGAUCCCUUUUAACCCGGGAUUCUGCACCACCAUC------GGCCUCCGCUCCUCUCGUCUCCGCCAGUCGAGUAUUAAGUAAAUGUU-GGGGCC--UAGGA .(((.(((((((((.........))))))))).))).((...------((((((.((....((((.((.....)).))))............)).-))))))--..)). ( -35.39) >DroEre_CAF1 14780 105 + 1 GGGUCCAGGAUCCCUUUUAACCCGGGAUUCUGCACCACCUCUUGCUCUGGGC-UCCUUCCCCUCGACUCCUCCAGUCGAGUAUUGAGUAAAUGUU-GGGGCC--UAGGA .(((.(((((((((.........))))))))).))).........(((((((-(((.....(((((((.....)))))))((((.....))))..-))))))--)))). ( -43.60) >DroYak_CAF1 26154 93 + 1 GGGUCCAGGAUCCCUUUUAACCCGGGAUUCUGCACCACCUCC-------------ACUUCGCUCGACUCCUCCACUCGAGUAUUGAGUAAAUGUU-GGGGCC--UAGGA .(((.(((((((((.........))))))))).))).((((.-------------((((..(((.((((........))))...)))..)).)).-))))..--..... ( -32.00) >consensus GGGUCCAGGAUCCCUUUUAACCCGGGAUUCUGCACCACCAUC______GGCCUCCACUCCUCUCGUCUCCGCCAGUCGAGUAUUAAGUAAAUGUU_GGGGCC__UAGGA .(((.(((((((((.........))))))))).)))....................((((.((((.((.....)).))))((((.....))))...))))......... (-22.18 = -22.50 + 0.32)

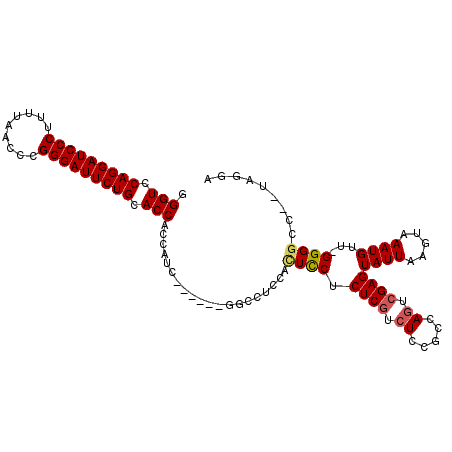
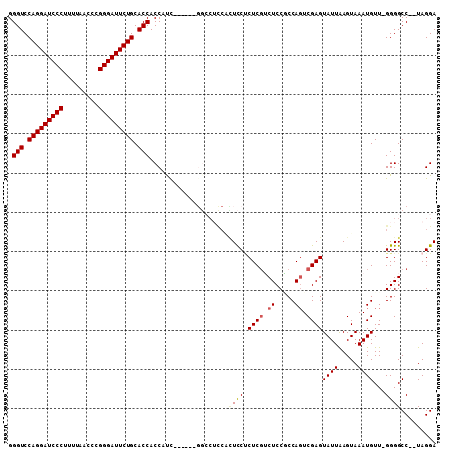
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:43 2006