
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,673,095 – 10,673,212 |
| Length | 117 |
| Max. P | 0.681724 |

| Location | 10,673,095 – 10,673,212 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -19.86 |
| Consensus MFE | -12.16 |
| Energy contribution | -11.96 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10673095 117 + 23771897 CAUGCAUUAAUCGAAAUUCCAAUAUUCCGCAACCCAUUAUUGCGGACCCUGCUGUGCUCGUCUAUCUACAAGGACAAACAAUCAUGUCCUACACUUGUUCCCAUACUCCCAGCAUCC .((((......(((.((..((....(((((((.......)))))))...))..))..)))..........((((((........)))))).....................)))).. ( -21.60) >DroSec_CAF1 7774 114 + 1 CUUGCAUUGAUCGGAAUCCCAAUAUUCCACAACCCAUUAUUGCGGACCCUGCUGUGCUCGU--AUCUACAAGGACAAACAAUCAUGUCCUACACUUGUUCCCAUACUC-CAGCAUCC ...(((......(((((......)))))....((((....)).))....))).(((((.((--((..(((((((((........)))))).....)))....))))..-.))))).. ( -21.00) >DroSim_CAF1 7675 114 + 1 CAUGCAUUAAUCGGAAUCCCAAUAUUCCACAACCCAUUAUUGCGGACCCUGCUGUGCUCGU--AUCUACAAGGACAAACAAUCAUGUCCUACACUUGUUCCCAUACUC-CAGCAUCC ...(((......(((((......)))))....((((....)).))....))).(((((.((--((..(((((((((........)))))).....)))....))))..-.))))).. ( -21.00) >DroEre_CAF1 8147 95 + 1 -------------------CAAUAUUCCACAACCCAUUAUUGCGAACCCAGCUGUGCUCGU--AUCUACAAGGACAAACAAUCAUGUCCUACACUUGUUCCCACACUC-CAGCAUCC -------------------(((((.............)))))........((((.(...((--....(((((((((........)))))).....)))....))...)-)))).... ( -12.42) >DroYak_CAF1 19104 114 + 1 CAUGUAUUAAUCCGCACAGCAAUAGCCCACAAUCCAUUAUUGCGAACCCCGCUGUGCUCGU--AUCUACAAGGACAAACAAUCAUGUCCUACGCUUGUUCCCAUACUC-CGGCAUCC ..((((.......(((((((.......(.((((.....)))).)......)))))))....--...))))((((((........))))))..(((.((......))..-.))).... ( -23.26) >consensus CAUGCAUUAAUCGGAAUCCCAAUAUUCCACAACCCAUUAUUGCGGACCCUGCUGUGCUCGU__AUCUACAAGGACAAACAAUCAUGUCCUACACUUGUUCCCAUACUC_CAGCAUCC ................................((((....)).)).....((((.....((......(((((((((........)))))).....)))......))...)))).... (-12.16 = -11.96 + -0.20)

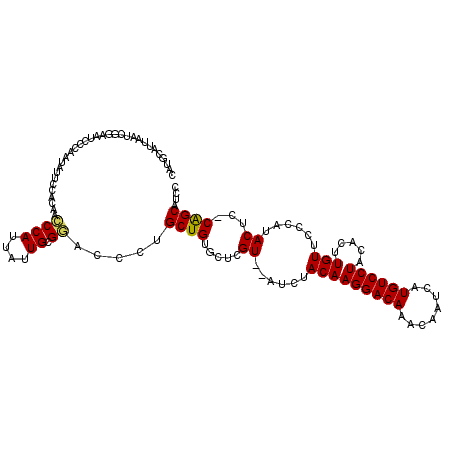
| Location | 10,673,095 – 10,673,212 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -32.26 |
| Consensus MFE | -23.02 |
| Energy contribution | -22.62 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681724 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10673095 117 - 23771897 GGAUGCUGGGAGUAUGGGAACAAGUGUAGGACAUGAUUGUUUGUCCUUGUAGAUAGACGAGCACAGCAGGGUCCGCAAUAAUGGGUUGCGGAAUAUUGGAAUUUCGAUUAAUGCAUG ..((((....(((.((..(....((((.(((((........)))))((((......))))))))..(((..((((((((.....))))))))...)))...)..)))))...)))). ( -31.60) >DroSec_CAF1 7774 114 - 1 GGAUGCUG-GAGUAUGGGAACAAGUGUAGGACAUGAUUGUUUGUCCUUGUAGAU--ACGAGCACAGCAGGGUCCGCAAUAAUGGGUUGUGGAAUAUUGGGAUUCCGAUCAAUGCAAG ((((.(((-.....((....)).((((.(((((........)))))((((....--))))))))..))).)))).......((.((((((((((......))))))..)))).)).. ( -32.70) >DroSim_CAF1 7675 114 - 1 GGAUGCUG-GAGUAUGGGAACAAGUGUAGGACAUGAUUGUUUGUCCUUGUAGAU--ACGAGCACAGCAGGGUCCGCAAUAAUGGGUUGUGGAAUAUUGGGAUUCCGAUUAAUGCAUG ..((((..-.(((.(((((.(..((((.(((((........)))))((((....--))))))))..(((..((((((((.....))))))))...))).).))))))))...)))). ( -32.70) >DroEre_CAF1 8147 95 - 1 GGAUGCUG-GAGUGUGGGAACAAGUGUAGGACAUGAUUGUUUGUCCUUGUAGAU--ACGAGCACAGCUGGGUUCGCAAUAAUGGGUUGUGGAAUAUUG------------------- ....((((-(..((((...(((.....((((((........)))))))))...)--)))..).))))....((((((((.....))))))))......------------------- ( -24.60) >DroYak_CAF1 19104 114 - 1 GGAUGCCG-GAGUAUGGGAACAAGCGUAGGACAUGAUUGUUUGUCCUUGUAGAU--ACGAGCACAGCGGGGUUCGCAAUAAUGGAUUGUGGGCUAUUGCUGUGCGGAUUAAUACAUG .(((.((.-..(((((....)..(((.((((((........)))))))))..))--))..(((((((((((((((((((.....)))))))))).))))))))))))))........ ( -39.70) >consensus GGAUGCUG_GAGUAUGGGAACAAGUGUAGGACAUGAUUGUUUGUCCUUGUAGAU__ACGAGCACAGCAGGGUCCGCAAUAAUGGGUUGUGGAAUAUUGGGAUUCCGAUUAAUGCAUG ((((.(((......((....)).((((.(((((........)))))((((......))))))))..))).))))(((((.....)))))............................ (-23.02 = -22.62 + -0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:36 2006