| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,672,152 – 10,672,248 |
| Length | 96 |
| Max. P | 0.768861 |

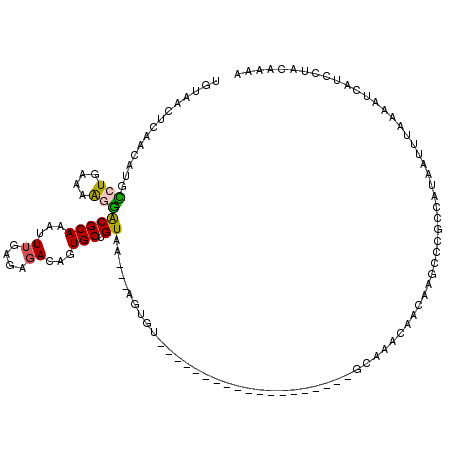
| Location | 10,672,152 – 10,672,248 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.88 |
| Mean single sequence MFE | -24.56 |
| Consensus MFE | -3.32 |
| Energy contribution | -4.36 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.14 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10672152 96 + 23771897 UGUUGUAGUAUGAUUUCAAAUUAUGGCGGGCUUGUUGUUUGC-------------------ACACU---UUACAGCACUAUCUCUCAAAUUUGCGUCCU-UGCAGGCA-AUUGAGUUACA ((((((((((((((.....))))))((((((.....))))))-------------------.....---))))))))......(((((.((((((....-))))))..-.)))))..... ( -21.80) >DroSec_CAF1 6817 120 + 1 UUUUGUAGAAUGAUUUUAAAUUAUGGCGGGCUUGUUGAUUUGAUUUGAGAGACAGUGCUGUAAAGUCUGCUACAGCACUGUCUCUCAAAUUUGCGUCCUUUUCAGGCAUGUUGAGUUACA ...(((((.(((((.....)))))((((.(((((..(((..(((((((((((((((((((((........)))))))))))))))))))))...))).....))))).))))...))))) ( -45.60) >DroSim_CAF1 6734 98 + 1 UUUUGUAGAAUAAUUUUAAAUUAUGGCGGGCUUGUUGGUUGC-------------------ACACU---UUACAGCACUGUCUCUCAAAUUUGCGUCCUUUUCAGGCAUGUUGAGUUACA ...(((((................((((((((.((.(((...-------------------..)))---..))))).))))).(((((...(((.(.......).)))..)))))))))) ( -19.10) >DroEre_CAF1 7311 85 + 1 GUUUGUAAGAUGCUUUUAAAUUAUGGCGGGCUUUUUGUUUGC-------------------ACACU---UUACAGCACUUUCUCUCAAAUUUGCGCAUCUUUUAGCC------------- (((...(((((((..........((((((((.....))))))-------------------.))..---.....(((..((......))..))))))))))..))).------------- ( -18.80) >DroYak_CAF1 18144 95 + 1 GUUUGUUUGGUGCUUUUAAAUUAUGGCGGGCU---UGUUUGC-------------------ACACU---UUACAGCACUAUAUCUCAAAUUUGCGUCGUUCUCAAACAUUUUGAGUGAGA (((((..(((((((..((((...(((((((..---..)))))-------------------.)).)---))).))))))).....))))).....((...(((((.....)))))...)) ( -17.50) >consensus UUUUGUAGAAUGAUUUUAAAUUAUGGCGGGCUUGUUGUUUGC___________________ACACU___UUACAGCACUAUCUCUCAAAUUUGCGUCCUUUUCAGGCAUGUUGAGUUACA ...(((((.(((((.....))))).((((((.....))))))...........................)))))(((..............))).......................... ( -3.32 = -4.36 + 1.04)

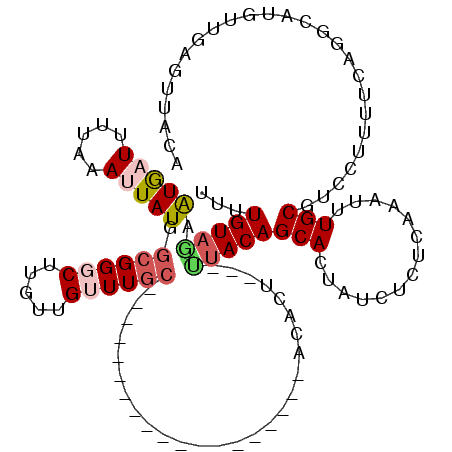
| Location | 10,672,152 – 10,672,248 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.88 |
| Mean single sequence MFE | -19.62 |
| Consensus MFE | -3.32 |
| Energy contribution | -3.16 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.17 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10672152 96 - 23771897 UGUAACUCAAU-UGCCUGCA-AGGACGCAAAUUUGAGAGAUAGUGCUGUAA---AGUGU-------------------GCAAACAACAAGCCCGCCAUAAUUUGAAAUCAUACUACAACA ((((.(((((.-....(((.-.....)))...))))).(((.(.(((....---..(((-------------------....)))...))).)..((.....))..)))....))))... ( -12.60) >DroSec_CAF1 6817 120 - 1 UGUAACUCAACAUGCCUGAAAAGGACGCAAAUUUGAGAGACAGUGCUGUAGCAGACUUUACAGCACUGUCUCUCAAAUCAAAUCAACAAGCCCGCCAUAAUUUAAAAUCAUUCUACAAAA ((((..........(((....)))......(((((((((((((((((((((......)))))))))))))))))))))...................................))))... ( -39.20) >DroSim_CAF1 6734 98 - 1 UGUAACUCAACAUGCCUGAAAAGGACGCAAAUUUGAGAGACAGUGCUGUAA---AGUGU-------------------GCAACCAACAAGCCCGCCAUAAUUUAAAAUUAUUCUACAAAA (((..(((((..(((((....))).)).....)))))..)))....((((.---.(((.-------------------((.........)).))).(((((.....)))))..))))... ( -13.30) >DroEre_CAF1 7311 85 - 1 -------------GGCUAAAAGAUGCGCAAAUUUGAGAGAAAGUGCUGUAA---AGUGU-------------------GCAAACAAAAAGCCCGCCAUAAUUUAAAAGCAUCUUACAAAC -------------......((((((((((..(((....)))..))).....---.(((.-------------------((.........)).)))............)))))))...... ( -18.10) >DroYak_CAF1 18144 95 - 1 UCUCACUCAAAAUGUUUGAGAACGACGCAAAUUUGAGAUAUAGUGCUGUAA---AGUGU-------------------GCAAACA---AGCCCGCCAUAAUUUAAAAGCACCAAACAAAC .....(((((...(((((.(.....).)))))))))).....(((((.(((---((((.-------------------((.....---.)).))).....))))..)))))......... ( -14.90) >consensus UGUAACUCAACAUGCCUGAAAAGGACGCAAAUUUGAGAGACAGUGCUGUAA___AGUGU___________________GCAAACAACAAGCCCGCCAUAAUUUAAAAUCAUCCUACAAAA ..............(((....)))(((((...((....))...))).))....................................................................... ( -3.32 = -3.16 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:33 2006