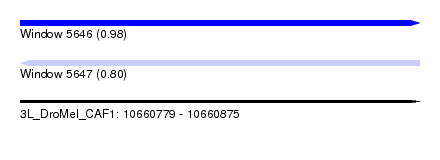
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,660,779 – 10,660,875 |
| Length | 96 |
| Max. P | 0.983728 |

| Location | 10,660,779 – 10,660,875 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -25.71 |
| Consensus MFE | -21.14 |
| Energy contribution | -20.82 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10660779 96 + 23771897 CCCAACACUACUCACCUGUCAAAAUACGCCGAAGUCGGCCUCUGUAUGAGGAUUUAUCAAAAACCGAGGUGACUUCGUGCAAGUGGCACUUAAUAU ................(((((......((((((((((.((((.((.(((.......)))...)).)))))))))))).))...)))))........ ( -25.40) >DroSec_CAF1 5428 91 + 1 CCCAACAUUGC-----UGUCAAAAUAAACCGAAGUCGGCCUCUGUAUGAGGAUUUAUCAAAAACCGAGGUGACUUCGUGCAAGUGGCACUAAAUAU .(((...((((-----.((........))((((((((.((((.((.(((.......)))...)).)))))))))))).)))).))).......... ( -23.40) >DroSim_CAF1 4872 96 + 1 CCCAACACUACUCACCUGUCAAAAUAAACCGAAGUCGGCCUCUGUAUGAGGAUUUAUCAAAAACCGAGGUGACUUCGUGCAAGUGGCACUAAAUAU ................(((((........((((((((.((((.((.(((.......)))...)).))))))))))))......)))))........ ( -22.54) >DroEre_CAF1 5066 95 + 1 CCCAACACUACUCACCUGUCAA-AUCAGCCGAAGUCGGCCUCUGUAUGAGGAUUUUCCAAAAACCGAGGAGACUUCGUGCAAGUGGCACUAAAGAU ................(((((.-....(((((((((..((((.((.(..((.....))..).)).)))).))))))).))...)))))........ ( -27.00) >DroYak_CAF1 4942 95 + 1 CCCAACACUACUCACCUGUCAA-AUCAGCCGAAGUCGGCCUCUGUAUGAGGAUUUUCCAAAAACCGAGGAGACUUCGUGCGAGUGGCACUAAAUAU .......((((((...((....-..))(((((((((..((((.((.(..((.....))..).)).)))).))))))).)))))))).......... ( -30.20) >consensus CCCAACACUACUCACCUGUCAAAAUAAGCCGAAGUCGGCCUCUGUAUGAGGAUUUAUCAAAAACCGAGGUGACUUCGUGCAAGUGGCACUAAAUAU ................(((((......(((((((((..((((.....))))....(((.........)))))))))).))...)))))........ (-21.14 = -20.82 + -0.32)

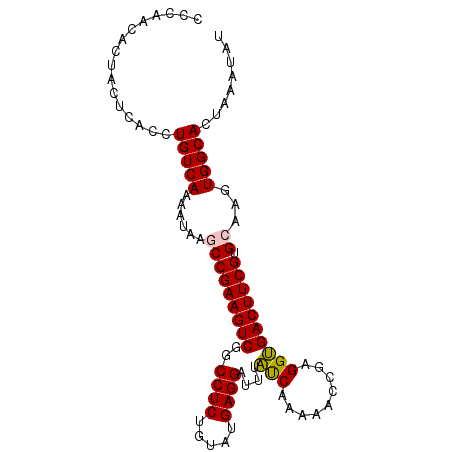
| Location | 10,660,779 – 10,660,875 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.66 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -22.54 |
| Energy contribution | -23.34 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10660779 96 - 23771897 AUAUUAAGUGCCACUUGCACGAAGUCACCUCGGUUUUUGAUAAAUCCUCAUACAGAGGCCGACUUCGGCGUAUUUUGACAGGUGAGUAGUGUUGGG ((((((..((((....((.(((((((.((((.((...(((.......))).)).))))..)))))))))((......)).))))..)))))).... ( -28.30) >DroSec_CAF1 5428 91 - 1 AUAUUUAGUGCCACUUGCACGAAGUCACCUCGGUUUUUGAUAAAUCCUCAUACAGAGGCCGACUUCGGUUUAUUUUGACA-----GCAAUGUUGGG ..........((((((((.(((((((.((((.((...(((.......))).)).))))..)))))))(((......))).-----)))).).))). ( -26.20) >DroSim_CAF1 4872 96 - 1 AUAUUUAGUGCCACUUGCACGAAGUCACCUCGGUUUUUGAUAAAUCCUCAUACAGAGGCCGACUUCGGUUUAUUUUGACAGGUGAGUAGUGUUGGG ....((((..(.(((..(.(((((((.((((.((...(((.......))).)).))))..)))))))(((......)))..)..))).)..)))). ( -29.20) >DroEre_CAF1 5066 95 - 1 AUCUUUAGUGCCACUUGCACGAAGUCUCCUCGGUUUUUGGAAAAUCCUCAUACAGAGGCCGACUUCGGCUGAU-UUGACAGGUGAGUAGUGUUGGG ....((((..(.(((..(.(((((((.((((.((....((.....))....)).))))..))))))).(((..-....))))..))).)..)))). ( -31.50) >DroYak_CAF1 4942 95 - 1 AUAUUUAGUGCCACUCGCACGAAGUCUCCUCGGUUUUUGGAAAAUCCUCAUACAGAGGCCGACUUCGGCUGAU-UUGACAGGUGAGUAGUGUUGGG ....((((..(.((((((.(((((((.((((.((....((.....))....)).))))..))))))).(((..-....))))))))).)..)))). ( -33.90) >consensus AUAUUUAGUGCCACUUGCACGAAGUCACCUCGGUUUUUGAUAAAUCCUCAUACAGAGGCCGACUUCGGCUUAUUUUGACAGGUGAGUAGUGUUGGG .....((.((((((((((((((((((.((((.((...(((.......))).)).))))..)))))))........)).)))))).))).))..... (-22.54 = -23.34 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:29 2006