| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 1,047,098 – 1,047,217 |
| Length | 119 |
| Max. P | 0.999530 |

| Location | 1,047,098 – 1,047,217 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -22.71 |
| Energy contribution | -23.31 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985505 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
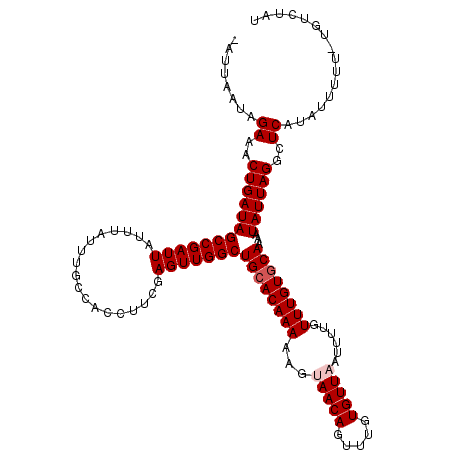
>3L_DroMel_CAF1 1047098 119 + 23771897 AAUUAAUAGAAACUGAUAGCCGAUUAUUUAUUUGCCACCUUCGAGUUGGCUGCACAAAAAGUAACAGUUUGUGUUAAUAUUGUUUGUGCAAAAUAUUAGGCUCAUAUUUUUUUGUCUAU .....(((((((...((((((...(((((....((((.(.....).))))((((((((...(((((.....)))))......)))))))))))))...)))).))...))))))).... ( -27.40) >DroSec_CAF1 21650 110 + 1 -AUUAAUAGAAACUGAUAGCCGAUUAUUUAUUUGCCACCUUCGAGUUGGCUGCACAAAAAGUAACAGUUUGUGUUCAUUUUGUUUGUGCAAAAUAUUAGGCUCAUAUUUUU-------- -.......((..(((((((((((((..................)))))))((((((((((((((((.....)))).))))..))))))))...))))))..))........-------- ( -23.87) >DroSim_CAF1 22412 110 + 1 -AUUAAUAGAAACUGAUAGCCGAUUAUUUAUUUGCCACCUUCGAGUUGGCUGCACAAAAAGUAACAGUUUGUGUUCAUUUUGUUUGUGCAAAAUAUUAGGCUCAUAUUUUU-------- -.......((..(((((((((((((..................)))))))((((((((((((((((.....)))).))))..))))))))...))))))..))........-------- ( -23.87) >DroEre_CAF1 24705 117 + 1 -AUUAAUAGAAACUGAUAGCCGAUUAUUUAUUUGCCACCUUCGAGUUGGCUGCACAAAAAGUAACAGUUUGUGUUAAUUUUGUUUGUACAAAAUAUUAGGCUCAUAUUUU-GUGUCUAU -..(((((.((((((.(((((((((..................))))))))).((.....))..)))))).)))))(((((((....)))))))..(((((.((.....)-).))))). ( -26.97) >DroYak_CAF1 22614 117 + 1 -AUUAAUAGAAACUGAUAGCCGAUUAUUUACUUGCCACCUUCGAGUUGGCUGCACAAAAAGUAACAGUUUGUGUUAAUAUUGUUUGUGCAAAUUAUUAGGCUCAUAUUUG-UUGUCUAU -....((((((((.(((((((............((((.(.....).))))((((((((...(((((.....)))))......))))))))........))))...))).)-)).))))) ( -27.20) >consensus _AUUAAUAGAAACUGAUAGCCGAUUAUUUAUUUGCCACCUUCGAGUUGGCUGCACAAAAAGUAACAGUUUGUGUUAAUUUUGUUUGUGCAAAAUAUUAGGCUCAUAUUUUU_UGUCUAU ........((..(((((((((((((..................)))))))((((((((...(((((.....)))))......))))))))...))))))..))................ (-22.71 = -23.31 + 0.60)

| Location | 1,047,098 – 1,047,217 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 92.51 |
| Mean single sequence MFE | -22.60 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
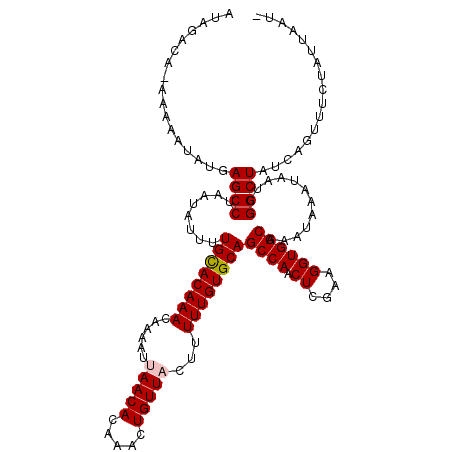
>3L_DroMel_CAF1 1047098 119 - 23771897 AUAGACAAAAAAAUAUGAGCCUAAUAUUUUGCACAAACAAUAUUAACACAAACUGUUACUUUUUGUGCAGCCAACUCGAAGGUGGCAAAUAAAUAAUCGGCUAUCAGUUUCUAUUAAUU (((((((........))((((........((((((((......(((((.....)))))...))))))))((((.((....))))))............)))).......)))))..... ( -22.60) >DroSec_CAF1 21650 110 - 1 --------AAAAAUAUGAGCCUAAUAUUUUGCACAAACAAAAUGAACACAAACUGUUACUUUUUGUGCAGCCAACUCGAAGGUGGCAAAUAAAUAAUCGGCUAUCAGUUUCUAUUAAU- --------.........((((........((((((((.......((((.....))))....))))))))((((.((....))))))............))))................- ( -21.70) >DroSim_CAF1 22412 110 - 1 --------AAAAAUAUGAGCCUAAUAUUUUGCACAAACAAAAUGAACACAAACUGUUACUUUUUGUGCAGCCAACUCGAAGGUGGCAAAUAAAUAAUCGGCUAUCAGUUUCUAUUAAU- --------.........((((........((((((((.......((((.....))))....))))))))((((.((....))))))............))))................- ( -21.70) >DroEre_CAF1 24705 117 - 1 AUAGACAC-AAAAUAUGAGCCUAAUAUUUUGUACAAACAAAAUUAACACAAACUGUUACUUUUUGUGCAGCCAACUCGAAGGUGGCAAAUAAAUAAUCGGCUAUCAGUUUCUAUUAAU- (((((.((-........((((........((((((((...((.(((((.....))))).))))))))))((((.((....))))))............))))....)).)))))....- ( -22.50) >DroYak_CAF1 22614 117 - 1 AUAGACAA-CAAAUAUGAGCCUAAUAAUUUGCACAAACAAUAUUAACACAAACUGUUACUUUUUGUGCAGCCAACUCGAAGGUGGCAAGUAAAUAAUCGGCUAUCAGUUUCUAUUAAU- (((((.((-(.......((((........((((((((......(((((.....)))))...))))))))((((.((....))))))............))))....))))))))....- ( -24.50) >consensus AUAGACA_AAAAAUAUGAGCCUAAUAUUUUGCACAAACAAAAUUAACACAAACUGUUACUUUUUGUGCAGCCAACUCGAAGGUGGCAAAUAAAUAAUCGGCUAUCAGUUUCUAUUAAU_ .................((((........((((((((......(((((.....)))))...))))))))((((.((....))))))............))))................. (-20.86 = -21.10 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:40:06 2006