
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,650,047 – 10,650,196 |
| Length | 149 |
| Max. P | 0.999992 |

| Location | 10,650,047 – 10,650,142 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -19.70 |
| Energy contribution | -20.10 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10650047 95 + 23771897 UAAGAACAAGCGCAUAAAAAAAAAAGAAUUGUGCCGCACAUAGAAGGCAGCACAACUAAUACUAGUUUUAGUGGCAGCCACUAGUGCUUGGACUA ..((..(((((((...............((((((.((.(......))).)))))).............((((((...)))))))))))))..)). ( -28.20) >DroSec_CAF1 12400 94 + 1 UAAGAACAAGCGCAUAAAAA-AAAGGAAUUGUGCCGCACAUAGAAGGCAACCCAACUAAUACUAGUUUUAGUGGCAGCCACUAGUGCUUGGACUA ..((..(((((((.......-...((.....((((..........))))..))(((((....))))).((((((...)))))))))))))..)). ( -25.90) >DroSim_CAF1 11766 93 + 1 UAAGAACAAGCGCAUAAAA--AAAGGAAUUGUGCCGCACAUAGAAGGCAGCACAACUAAUACUAGUUUUAGUGGCAGCCACUAGUGCUCGGACUA ..((..(.(((((......--.......((((((.((.(......))).)))))).............((((((...))))))))))).)..)). ( -24.10) >DroEre_CAF1 12851 94 + 1 UAAGAACAAGCGCAUAAAAA-AAAGGAAUUGUGCCGCACAUAGAAGGCAGCACAACUAAUACUAGUUUUAGUAGCAGCCACUAGUGGUUGUAUUA ....................-........(((((.((.(......))).)))))(((((........))))).(((((((....))))))).... ( -20.90) >DroYak_CAF1 12744 94 + 1 UAAGAACAAGCGCAUAAAAA-AAAGGAAUUGUGCCGCACAUAGAAGGCAGCACAACUAAUACUAGUUUUAGUAGCAGCCACUAGUGCUGGUAUUU ....................-.......((((((.((.(......))).))))))..(((((((((.(((((.......))))).))))))))). ( -21.50) >consensus UAAGAACAAGCGCAUAAAAA_AAAGGAAUUGUGCCGCACAUAGAAGGCAGCACAACUAAUACUAGUUUUAGUGGCAGCCACUAGUGCUUGGACUA .........(((((...............))))).((((.(((..(((.((...(((((........))))).)).))).)))))))........ (-19.70 = -20.10 + 0.40)



| Location | 10,650,047 – 10,650,142 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 94.39 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -24.26 |
| Energy contribution | -24.34 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.93 |
| SVM decision value | 5.10 |
| SVM RNA-class probability | 0.999973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10650047 95 - 23771897 UAGUCCAAGCACUAGUGGCUGCCACUAAAACUAGUAUUAGUUGUGCUGCCUUCUAUGUGCGGCACAAUUCUUUUUUUUUUAUGCGCUUGUUCUUA .((..(((((..((((((...))))))......((((.((((((((((((......).)))))))))))...........)))))))))..)).. ( -30.00) >DroSec_CAF1 12400 94 - 1 UAGUCCAAGCACUAGUGGCUGCCACUAAAACUAGUAUUAGUUGGGUUGCCUUCUAUGUGCGGCACAAUUCCUUU-UUUUUAUGCGCUUGUUCUUA .((..(((((..((((((...))))))......((((.((..(((.((((..........)))).....)))..-))...)))))))))..)).. ( -22.90) >DroSim_CAF1 11766 93 - 1 UAGUCCGAGCACUAGUGGCUGCCACUAAAACUAGUAUUAGUUGUGCUGCCUUCUAUGUGCGGCACAAUUCCUUU--UUUUAUGCGCUUGUUCUUA .((..(((((..((((((...))))))......((((.((((((((((((......).))))))))))).....--....)))))))))..)).. ( -29.90) >DroEre_CAF1 12851 94 - 1 UAAUACAACCACUAGUGGCUGCUACUAAAACUAGUAUUAGUUGUGCUGCCUUCUAUGUGCGGCACAAUUCCUUU-UUUUUAUGCGCUUGUUCUUA ..........((.((((((((.(((((....))))).))))(((((((((......).))))))))........-........)))).))..... ( -22.90) >DroYak_CAF1 12744 94 - 1 AAAUACCAGCACUAGUGGCUGCUACUAAAACUAGUAUUAGUUGUGCUGCCUUCUAUGUGCGGCACAAUUCCUUU-UUUUUAUGCGCUUGUUCUUA ....((.(((((((((.............))))))...((((((((((((......).))))))))))).....-.........))).))..... ( -24.32) >consensus UAGUCCAAGCACUAGUGGCUGCCACUAAAACUAGUAUUAGUUGUGCUGCCUUCUAUGUGCGGCACAAUUCCUUU_UUUUUAUGCGCUUGUUCUUA .....(((((..((((((...))))))......((((.((((((((((((......).)))))))))))...........)))))))))...... (-24.26 = -24.34 + 0.08)


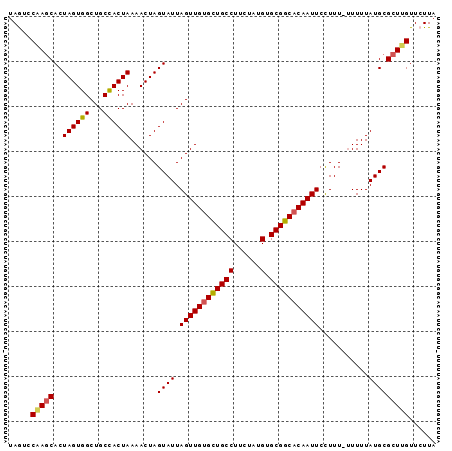
| Location | 10,650,104 – 10,650,196 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 94.46 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.18 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.71 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10650104 92 + 23771897 AAUACUAGUUUUAGUGGCAGCCACUAGUGCUUGGACUAGUGGCUGUAGUUGCAGCAUCACUUAGCUCGUUACCGUUAUUUGGUUGCAUACAG ..(((((....)))))((((((((((((......)))))))))))).((((((((..((..((((........))))..)))))))).)).. ( -32.70) >DroSec_CAF1 12456 92 + 1 AAUACUAGUUUUAGUGGCAGCCACUAGUGCUUGGACUAGUGGCUGUAGUUGCAGCAUCACUUAGCUCGUUACCGUUAUUUGGCUGCAUCCAG ..(((((....)))))((((((((((((......))))))))))))...((((((..((..((((........))))..))))))))..... ( -34.20) >DroSim_CAF1 11821 92 + 1 AAUACUAGUUUUAGUGGCAGCCACUAGUGCUCGGACUAGUGGCUGUAGUUGCAGCAUCACUUAGCUCGUUACCGUUAUUUGGCUGCAUCCAG ..(((((....)))))((((((((((((......))))))))))))...((((((..((..((((........))))..))))))))..... ( -34.20) >DroEre_CAF1 12907 92 + 1 AAUACUAGUUUUAGUAGCAGCCACUAGUGGUUGUAUUAGUCGCUGUAGUUGCAGCAUCACUUAGCUCGUUACCGUUAUUUAGCUGCAUCCAG ..(((((....)))))(((((.(((((((....))))))).)))))...((((((......((((........))))....))))))..... ( -24.60) >DroYak_CAF1 12800 92 + 1 AAUACUAGUUUUAGUAGCAGCCACUAGUGCUGGUAUUUGUGGCUGUAGUUGCAGCAUCACUUAGCUCGUUACCGUUAUUUAGCUGCAUCCAG ..(((((....)))))((((((((.((((....)))).))))))))...((((((......((((........))))....))))))..... ( -27.80) >consensus AAUACUAGUUUUAGUGGCAGCCACUAGUGCUUGGACUAGUGGCUGUAGUUGCAGCAUCACUUAGCUCGUUACCGUUAUUUGGCUGCAUCCAG ..(((((....)))))((((((((((((......))))))))))))...((((((......((((........))))....))))))..... (-30.42 = -30.18 + -0.24)

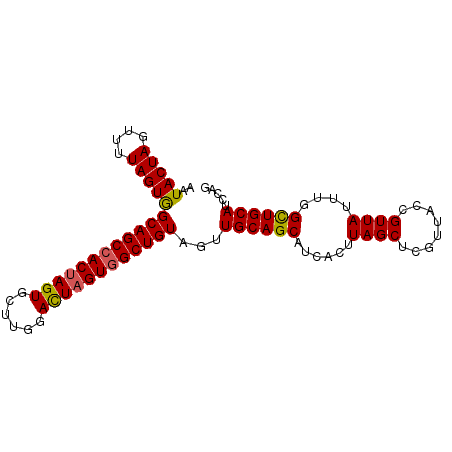

| Location | 10,650,104 – 10,650,196 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 94.46 |
| Mean single sequence MFE | -26.97 |
| Consensus MFE | -24.34 |
| Energy contribution | -25.54 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10650104 92 - 23771897 CUGUAUGCAACCAAAUAACGGUAACGAGCUAAGUGAUGCUGCAACUACAGCCACUAGUCCAAGCACUAGUGGCUGCCACUAAAACUAGUAUU .....(((.(((.......)))....(((........))))))....(((((((((((......)))))))))))..((((....))))... ( -26.80) >DroSec_CAF1 12456 92 - 1 CUGGAUGCAGCCAAAUAACGGUAACGAGCUAAGUGAUGCUGCAACUACAGCCACUAGUCCAAGCACUAGUGGCUGCCACUAAAACUAGUAUU ((((.((((((...((...(((.....)))..))...))))))....(((((((((((......))))))))))).........)))).... ( -30.60) >DroSim_CAF1 11821 92 - 1 CUGGAUGCAGCCAAAUAACGGUAACGAGCUAAGUGAUGCUGCAACUACAGCCACUAGUCCGAGCACUAGUGGCUGCCACUAAAACUAGUAUU ((((.((((((...((...(((.....)))..))...))))))....(((((((((((......))))))))))).........)))).... ( -30.60) >DroEre_CAF1 12907 92 - 1 CUGGAUGCAGCUAAAUAACGGUAACGAGCUAAGUGAUGCUGCAACUACAGCGACUAAUACAACCACUAGUGGCUGCUACUAAAACUAGUAUU .(((.((((((...((...(((.....)))..))...)))))).)))((((.((((..........)))).)))).(((((....))))).. ( -22.60) >DroYak_CAF1 12800 92 - 1 CUGGAUGCAGCUAAAUAACGGUAACGAGCUAAGUGAUGCUGCAACUACAGCCACAAAUACCAGCACUAGUGGCUGCUACUAAAACUAGUAUU .(((.((((((...((...(((.....)))..))...)))))).)))(((((((..............))))))).(((((....))))).. ( -24.24) >consensus CUGGAUGCAGCCAAAUAACGGUAACGAGCUAAGUGAUGCUGCAACUACAGCCACUAGUCCAAGCACUAGUGGCUGCCACUAAAACUAGUAUU ((((.((((((...((...(((.....)))..))...))))))....(((((((((((......))))))))))).........)))).... (-24.34 = -25.54 + 1.20)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:23 2006