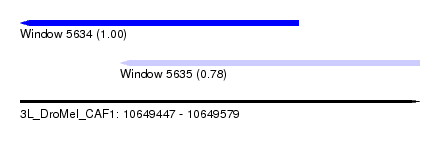
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,649,447 – 10,649,579 |
| Length | 132 |
| Max. P | 0.998431 |

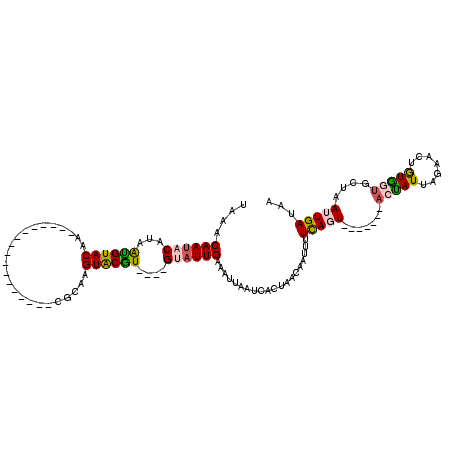
| Location | 10,649,447 – 10,649,539 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.94 |
| Mean single sequence MFE | -20.06 |
| Consensus MFE | -8.16 |
| Energy contribution | -8.24 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.41 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10649447 92 - 23771897 UAAACAAUACAUAGCGUACAC-----------------CGCAAGUACGU----GUAUUGAAAUUAAUCACUAACAAUUUUCAGU-------ACUAUUAGAACUGUGGUGCUAAUUGAUAA ....((((((...((((((..-----------------.....))))))----))))))..............(((((...(((-------(((((.......))))))))))))).... ( -21.30) >DroSec_CAF1 11820 92 - 1 UAAACAAUACAUAAUGUACAA-----------------CGCAAGUACGU----GCAUUGAAAUUAAUCACUAACAAUUUUCAUU-------ACUAUUAGAACUGUGCUGCUAAUUGAUAA ...........((((((((.(-----------------(....))..))----))))))..............(((((..((.(-------((..........))).))..))))).... ( -14.80) >DroSim_CAF1 11175 99 - 1 UAAACAAUACAUAAUGUACAA-----------------CGCAAGUACGU----GCAUUGAAAUUAAUCACUAACAAUUUUCAGUUUUCAGUUUCAUUAGAACUGUGCUGCUAAUUGAUAA ...........((((((((.(-----------------(....))..))----))))))..............(((((..((((...((((((.....)))))).))))..))))).... ( -23.60) >DroEre_CAF1 12240 109 - 1 UAAACAAUACAUAAUGUACAUACAGUACAGACACACACAUGUAGUGCGU----GUUUUGCAAUUAAUCACUAAACAUUUUUACU-------ACUAUCAAUGCUAUAGUGCUAAUUGAUAA ..............(((((.....)))))(((((.(((.....))).))----)))...((((((..(((((..((((..((..-------..))..))))...))))).)))))).... ( -20.20) >DroYak_CAF1 12114 108 - 1 UAAACAAUACCUAAUGUACAUAU-----GUAUACAUACGUCAAGUGCAUAUCUGUAUUGCAUUUAAUCACUAAACAUUUUCAGU-------ACUAUUAGUGCAAUAGUGCUAAUUGAUAA ....((((((...((((((..((-----(((....)))))...))))))....))))))......................(((-------((((((.....)))))))))......... ( -20.40) >consensus UAAACAAUACAUAAUGUACAA_________________CGCAAGUACGU____GUAUUGAAAUUAAUCACUAACAAUUUUCAGU_______ACUAUUAGAACUGUGGUGCUAAUUGAUAA ....((((((...((((((........................))))))....))))))....................(((((.......(((((.......)))))....)))))... ( -8.16 = -8.24 + 0.08)


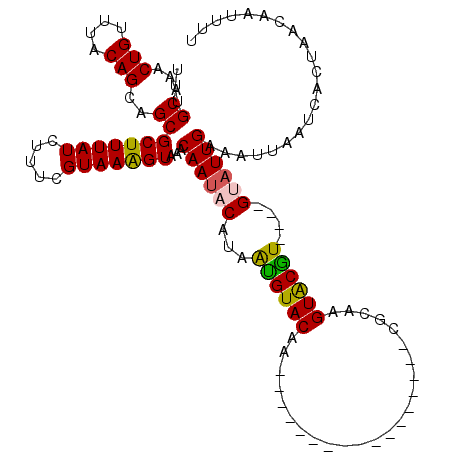
| Location | 10,649,480 – 10,649,579 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.44 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -14.90 |
| Energy contribution | -14.54 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10649480 99 - 23771897 UUUGCAAACUGUUUACAGCAGCGCUUUAUCUAUCGUAAAGUAAACAAUACAUAGCGUACAC-----------------CGCAAGUACGU----GUAUUGAAAUUAAUCACUAACAAUUUU ...((...(((....)))..))(((((((.....)))))))...((((((...((((((..-----------------.....))))))----))))))..................... ( -19.10) >DroSec_CAF1 11853 99 - 1 UUUGCAAACUGUUUACAGCAGCGCUUUAUCUUUCGUAAGGUAAACAAUACAUAAUGUACAA-----------------CGCAAGUACGU----GCAUUGAAAUUAAUCACUAACAAUUUU ...((...((((.....)))).))((((((((....)))))))).......((((((((.(-----------------(....))..))----))))))..................... ( -18.10) >DroSim_CAF1 11215 99 - 1 UUUGCAAACUGUUUACAGCAGCGCUUUAUCUUUCGUAAGGUAAACAAUACAUAAUGUACAA-----------------CGCAAGUACGU----GCAUUGAAAUUAAUCACUAACAAUUUU ...((...((((.....)))).))((((((((....)))))))).......((((((((.(-----------------(....))..))----))))))..................... ( -18.10) >DroEre_CAF1 12273 116 - 1 UUUGCAAACUGUUUACAGCAGCGCUUUAUCUUUCGUAAAGUAAACAAUACAUAAUGUACAUACAGUACAGACACACACAUGUAGUGCGU----GUUUUGCAAUUAAUCACUAAACAUUUU .((((((.((((.....)))).(((((((.....))))))).............(((((.....)))))(((((.(((.....))).))----))))))))).................. ( -24.10) >DroYak_CAF1 12147 115 - 1 UUUGCAAACUGUUUACAGCAGCGCUUUAUCUUUCGUAAAGUAAACAAUACCUAAUGUACAUAU-----GUAUACAUACGUCAAGUGCAUAUCUGUAUUGCAUUUAAUCACUAAACAUUUU ...((...(((....)))..))(((((((.....)))))))...((((((...((((((..((-----(((....)))))...))))))....))))))..................... ( -19.40) >consensus UUUGCAAACUGUUUACAGCAGCGCUUUAUCUUUCGUAAAGUAAACAAUACAUAAUGUACAA_________________CGCAAGUACGU____GUAUUGAAAUUAAUCACUAACAAUUUU ...((...(((....)))..))(((((((.....)))))))...((((((...((((((........................))))))....))))))..................... (-14.90 = -14.54 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:18 2006