
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
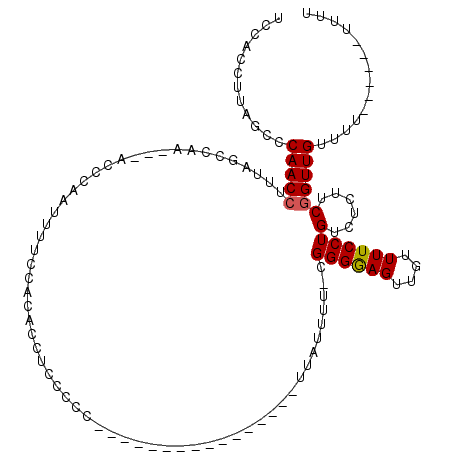
| Location | 10,643,212 – 10,643,306 |
| Length | 94 |
| Max. P | 0.996748 |

| Location | 10,643,212 – 10,643,306 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -14.52 |
| Consensus MFE | -8.06 |
| Energy contribution | -8.02 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.56 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
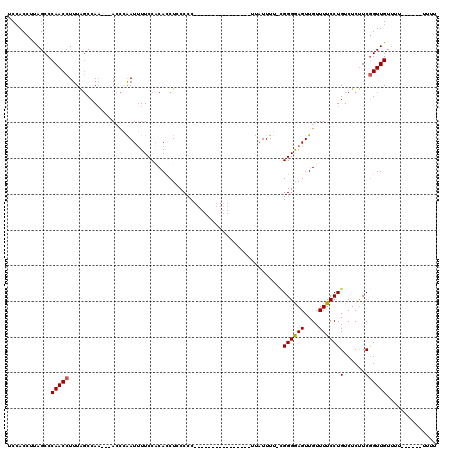
>3L_DroMel_CAF1 10643212 94 + 23771897 UCAACCUUAGCCCAACCUUUGGCAAA---ACCGAAUUUUCCACACCUCCCAC----------------UUAUUUU-CGGGGAGUUGUUUUCCUGUCUCUUCGGUUGUGUU------UUUU .(((((...(((........)))...---...((.......(((.(((((.(----------------.......-.)))))).))).......)).....)))))....------.... ( -20.24) >DroSec_CAF1 5708 96 + 1 UCCACCUUAGCCCAACCUUUAUCCAA---ACCAAAUUUUCCACACCUCCCCC----------------UUAUUUU-CGGGGAGUUGUUUUCCUGUCUCUUCGGUUGUUUUU----UUUUU ............(((((.........---............(((.((((((.----------------.......-.)))))).)))......(......)))))).....----..... ( -16.10) >DroSim_CAF1 5712 100 + 1 UCCACCUUAGCCCAACCUUUAGCCAA---AACGAAUUUUCCACACCUCCCCC----------------UUAUUUU-CGGGAAGUUGUUUUCCUGUCUCUUCGGUUGUUUUUUUUUUUUUU ............(((((...((.(((---(((.((((((((...........----------------.......-.))))))))))))...)).))....))))).............. ( -12.11) >DroEre_CAF1 5820 97 + 1 UCCACCUCAGCCCAACCUUUAGCCA----ACCCGAUUUUCCCCAUCUACCCC----------CAGCCCACAUUUUCUGGGAAGUUGUUUUCCUGUCUCCUCGGUUGUU---------UUU .......(((((.......(((.((----((..(((.......)))....((----------(((..........)))))..)))).....))).......)))))..---------... ( -12.44) >DroYak_CAF1 5746 110 + 1 UCCACCUUAGCCCAACCUUUAGCCAACCAACCCGAUGUUCCGCACCUACCCCACCCCCCUACCAUCCCAU-UUUGCAGGGGAGUUGUUUUCCUGUCUCUUCAGUUGUU---------UUC ............((((.....((....((......))....))............(((((..((......-..)).))))).)))).....(((......))).....---------... ( -11.70) >consensus UCCACCUUAGCCCAACCUUUAGCCAA___ACCCAAUUUUCCACACCUCCCCC________________UUAUUUU_CGGGGAGUUGUUUUCCUGUCUCUUCGGUUGUUUU______UUUU ............(((((............................................................((((((....))))))(......)))))).............. ( -8.06 = -8.02 + -0.04)

| Location | 10,643,212 – 10,643,306 |
|---|---|
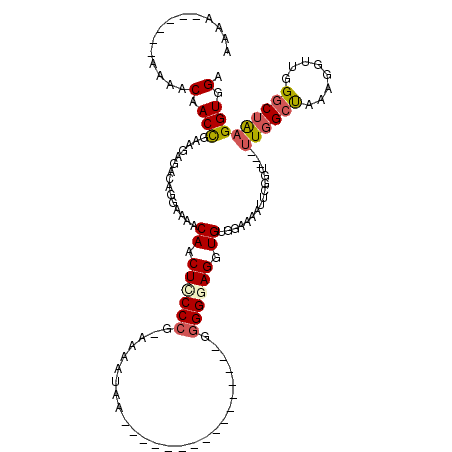
| Length | 94 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.12 |
| Mean single sequence MFE | -23.94 |
| Consensus MFE | -11.89 |
| Energy contribution | -12.53 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
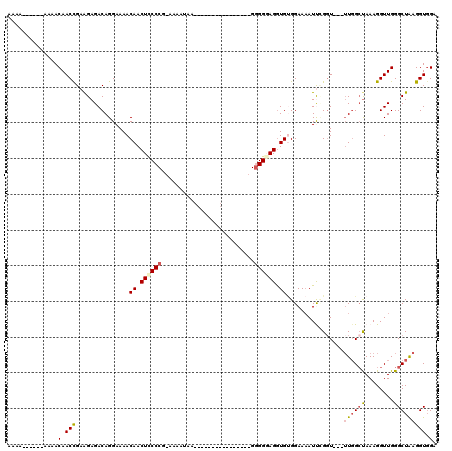
>3L_DroMel_CAF1 10643212 94 - 23771897 AAAA------AACACAACCGAAGAGACAGGAAAACAACUCCCCG-AAAAUAA----------------GUGGGAGGUGUGGAAAAUUCGGU---UUUGCCAAAGGUUGGGCUAAGGUUGA ....------....(((((...(((((.(((..(((.((((((.-.......----------------).))))).)))......))).))---)))(((........)))...))))). ( -25.20) >DroSec_CAF1 5708 96 - 1 AAAAA----AAAAACAACCGAAGAGACAGGAAAACAACUCCCCG-AAAAUAA----------------GGGGGAGGUGUGGAAAAUUUGGU---UUGGAUAAAGGUUGGGCUAAGGUGGA .....----.......(((..............(((.((((((.-.......----------------.)))))).))).......(((((---(..(.......)..)))))))))... ( -21.40) >DroSim_CAF1 5712 100 - 1 AAAAAAAAAAAAAACAACCGAAGAGACAGGAAAACAACUUCCCG-AAAAUAA----------------GGGGGAGGUGUGGAAAAUUCGUU---UUGGCUAAAGGUUGGGCUAAGGUGGA .................((.........))...(((.((((((.-.......----------------.)))))).)))......((((..---((((((........))))))..)))) ( -20.70) >DroEre_CAF1 5820 97 - 1 AAA---------AACAACCGAGGAGACAGGAAAACAACUUCCCAGAAAAUGUGGGCUG----------GGGGUAGAUGGGGAAAAUCGGGU----UGGCUAAAGGUUGGGCUGAGGUGGA ...---------..(((((..(....).......(((((((((((..........)))----------)))...(((.......))).)))----))......)))))............ ( -22.30) >DroYak_CAF1 5746 110 - 1 GAA---------AACAACUGAAGAGACAGGAAAACAACUCCCCUGCAAA-AUGGGAUGGUAGGGGGGUGGGGUAGGUGCGGAACAUCGGGUUGGUUGGCUAAAGGUUGGGCUAAGGUGGA ...---------..(((((..............((.((((((((((...-........))))))))))...)).((((.....)))).))))).((((((........))))))...... ( -30.10) >consensus AAAA______AAAACAACCGAAGAGACAGGAAAACAACUCCCCG_AAAAUAA________________GGGGGAGGUGUGGAAAAUUCGGU___UUGGCUAAAGGUUGGGCUAAGGUGGA ..............(.(((...............((.((((((..........................)))))).))................((((((........))))))))).). (-11.89 = -12.53 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:14 2006