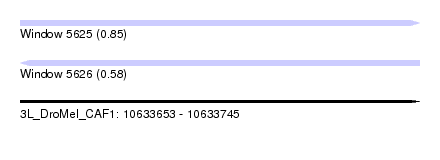
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,633,653 – 10,633,745 |
| Length | 92 |
| Max. P | 0.849125 |

| Location | 10,633,653 – 10,633,745 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 68.75 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -12.99 |
| Energy contribution | -14.49 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849125 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10633653 92 + 23771897 -AGCAGCUGUUGCACCUGUUGUC-------ACG-AAAAAAGGG-CCUAAUAAAUGG------GCUG---GUUACCCAAAAGGCAGCUGCAAAAGUUGCUAGGGAUAGUGGA -(((((((.(((((...((((((-------...-.......((-((((.....)))------)))(---(....))....))))))))))).)))))))............ ( -30.20) >DroSec_CAF1 515854 87 + 1 -----GCUGUUGCACCUGUUGUC-------ACG-AAUAAAGGG-UUUAAUAAAUGG------GCUGGUGGUUUCCCAAAAGGCAGCUGCAAAAGUUGCUAGGGAUAG---- -----(((((((.((((.((((.-------...-.)))).)))-).)))).....)------))........((((....(((((((.....))))))).))))...---- ( -21.00) >DroSim_CAF1 517770 78 + 1 -AGCAGCUGUUGCACCUGUUGUC-------ACG-AAAAA-----------UAAUGG------GCUG---GUUCCCCAAAAGGCAGCUGCAAAAGUUGCUAGGGAUAG---- -(((((((.(((((.((((((((-------.((-.....-----------...)))------))((---(....)))...))))).))))).)))))))........---- ( -23.30) >DroEre_CAF1 524148 89 + 1 -AGCAGCUGUUGCACCUGUUGUC-------ACG-AAACAAGGGACCUAAUUAAUGG------GCUG---GUUCCCCAAAGGGCAGCUGCUAAAGUUGCUAGGGACAG---- -(((((((...(((...((((((-------.(.-......((((((..........------...)---))))).....))))))))))...)))))))........---- ( -26.82) >DroWil_CAF1 810004 97 + 1 AGACGGCUCUUGCACCUGUUAGCCAAACACACACAACAAAGGG-AUUAAUAAAUUUUCUUUAAAUG---G--CCUCGACCGGCAGCUGCAAAAG--GCAAAAGAU------ ....(.((.(((((.(((((.................((((..-(.........)..))))....(---(--......))))))).))))).))--.).......------ ( -18.30) >DroYak_CAF1 526484 72 + 1 -AGCAGCUGUUGCACCUGUUGUC-------ACG-AAAAA-------UAAUAAAGAA------AAUG-------------GGGCAGCUGCAAGAGUUGCUGGGGAUAG---- -(((((((.(((((...((((((-------...-.....-------..........------....-------------.))))))))))).)))))))........---- ( -21.40) >consensus _AGCAGCUGUUGCACCUGUUGUC_______ACG_AAAAAAGGG___UAAUAAAUGG______GCUG___GUUCCCCAAAAGGCAGCUGCAAAAGUUGCUAGGGAUAG____ .(((((((.(((((.(((((............................................................))))).))))).)))))))............ (-12.99 = -14.49 + 1.50)

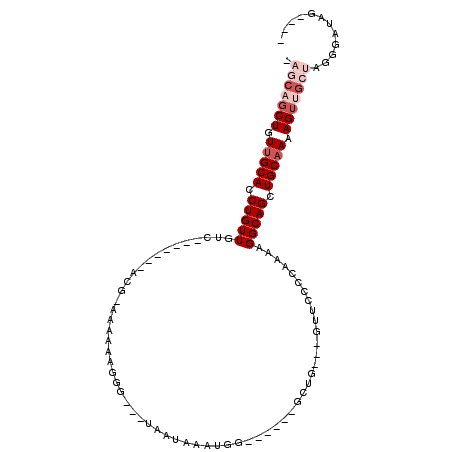
| Location | 10,633,653 – 10,633,745 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.75 |
| Mean single sequence MFE | -20.78 |
| Consensus MFE | -9.41 |
| Energy contribution | -10.67 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578232 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10633653 92 - 23771897 UCCACUAUCCCUAGCAACUUUUGCAGCUGCCUUUUGGGUAAC---CAGC------CCAUUUAUUAGG-CCCUUUUUU-CGU-------GACAACAGGUGCAACAGCUGCU- ............((((.((.(((((.((((((..(((((...---..))------)))......)))-)........-.((-------....)))).))))).)).))))- ( -25.50) >DroSec_CAF1 515854 87 - 1 ----CUAUCCCUAGCAACUUUUGCAGCUGCCUUUUGGGAAACCACCAGC------CCAUUUAUUAAA-CCCUUUAUU-CGU-------GACAACAGGUGCAACAGC----- ----.............((.(((((.(((.((..(((....)))..)).------.(((..((....-......)).-.))-------)....))).))))).)).----- ( -15.20) >DroSim_CAF1 517770 78 - 1 ----CUAUCCCUAGCAACUUUUGCAGCUGCCUUUUGGGGAAC---CAGC------CCAUUA-----------UUUUU-CGU-------GACAACAGGUGCAACAGCUGCU- ----........((((.((.(((((.(((....((((....)---))).------.(((..-----------.....-.))-------)....))).))))).)).))))- ( -20.50) >DroEre_CAF1 524148 89 - 1 ----CUGUCCCUAGCAACUUUAGCAGCUGCCCUUUGGGGAAC---CAGC------CCAUUAAUUAGGUCCCUUGUUU-CGU-------GACAACAGGUGCAACAGCUGCU- ----.................((((((((....((((....)---))).------...........(..(((((((.-...-------))))..)))..)..))))))))- ( -23.80) >DroWil_CAF1 810004 97 - 1 ------AUCUUUUGC--CUUUUGCAGCUGCCGGUCGAGG--C---CAUUUAAAGAAAAUUUAUUAAU-CCCUUUGUUGUGUGUGUUUGGCUAACAGGUGCAAGAGCCGUCU ------.......(.--((((((((.(((..((((((((--(---(((.(((((.............-..)))))..))).)).)))))))..))).)))))))).).... ( -25.46) >DroYak_CAF1 526484 72 - 1 ----CUAUCCCCAGCAACUCUUGCAGCUGCCC-------------CAUU------UUCUUUAUUA-------UUUUU-CGU-------GACAACAGGUGCAACAGCUGCU- ----........((((.((.(((((.(((...-------------(((.------..........-------.....-.))-------)....))).))))).)).))))- ( -14.23) >consensus ____CUAUCCCUAGCAACUUUUGCAGCUGCCUUUUGGGGAAC___CAGC______CCAUUUAUUA___CCCUUUUUU_CGU_______GACAACAGGUGCAACAGCUGCU_ .............(((.((.(((((.(((((.....)).........................................((........))..))).))))).)).))).. ( -9.41 = -10.67 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:02:10 2006