| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
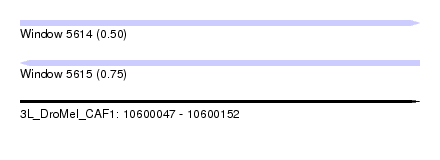
| Location | 10,600,047 – 10,600,152 |
| Length | 105 |
| Max. P | 0.747184 |

| Location | 10,600,047 – 10,600,152 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -27.35 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.16 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
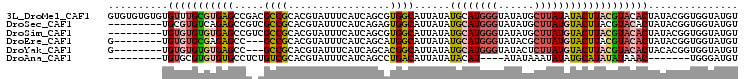
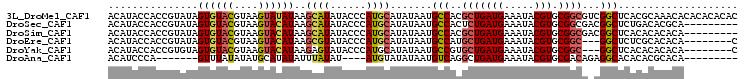
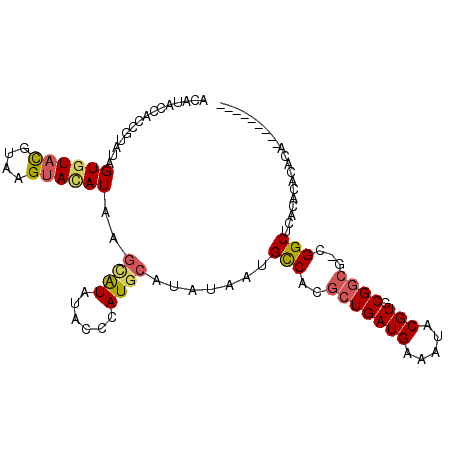
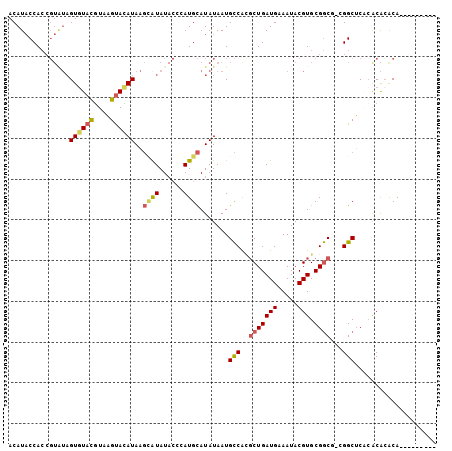
>3L_DroMel_CAF1 10600047 105 + 23771897 GUGUGUGUGUGUUUGCGUGAGCCGACGCCGCACGUAUUUCAUCAGCGUGGCAUUAUAUGCAUGGGUAUAUGCUUAUAUACUUACGUACACUAUACGGUGGUAUGU ..(((((.(((((.((....)).))))))))))((((....((((((((......))))).)))....))))..(((((((..((((.....))))..))))))) ( -35.60) >DroSec_CAF1 482437 96 + 1 ---------UGCGUGUCAGAGCCGUCGCCGCACGUAUUUCAUCAGAGUGGCAUUAUAUGCAUGGGUAUAUGCUUAUGUACUUACGUACACUAUACGGUGGUAUGU ---------.(((((.(...(((((.(((((.(...........).)))))......(((((((((....)))))))))..............))))).)))))) ( -24.90) >DroSim_CAF1 482497 96 + 1 ---------UGUGUGUGUGAGCCGUCGCCGCACGUAUUUCAUCAGCGUGGCAUUAUAUGCAUGGGUAUAUGCUUAUGUACUUACGUACACUAUACGGUGGUAUGU ---------.(((((((((((.....(((((.(...........).)))))......(((((((((....))))))))))))))))))))(((((....))))). ( -30.10) >DroEre_CAF1 489739 94 + 1 G--------UGUGUGCGAGAGCC---GCCGCACGUAUUUCAUCAGCAUGGCAUUAUAUGCAUGGGUAUACGCUUAUGUACUUACGUACACUAUACGGUGGUAUGU (--------((((.(((.....)---)))))))...........(((((.(((....(((((((((....)))))))))....((((.....))))))).))))) ( -27.50) >DroYak_CAF1 492234 94 + 1 G--------UGUGUGUGUGAGCC---GCCGCACGUAUUUCAUCAGCACGGCAUUAUAUGCAUGGGUAUACUCUUAUGUACUUACGUACACUACACGGUGGUAUGU (--------((((((((((.(((---(..((.............)).))))......((((((((......))))))))....))))))).)))).......... ( -25.22) >DroAna_CAF1 472698 85 + 1 ---------UGUGCGUGUGUGCCUCUGUCGCACGUAUUUCAUCAGCCUGACAUUAUAUACAU----AUAUAAAUAUAUGCAUAUAUAAAC-------UGGGAUGU ---------.(((((((((.((....)))))))))))..((((..((......(((((.(((----(((....)))))).))))).....-------.)))))). ( -20.80) >consensus _________UGUGUGUGUGAGCCG_CGCCGCACGUAUUUCAUCAGCGUGGCAUUAUAUGCAUGGGUAUAUGCUUAUGUACUUACGUACACUAUACGGUGGUAUGU ..........(((((((((((.....((((.................))))......((((((((......)))))))))))))))))))............... (-17.60 = -18.16 + 0.56)
| Location | 10,600,047 – 10,600,152 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.86 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -14.59 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.747184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
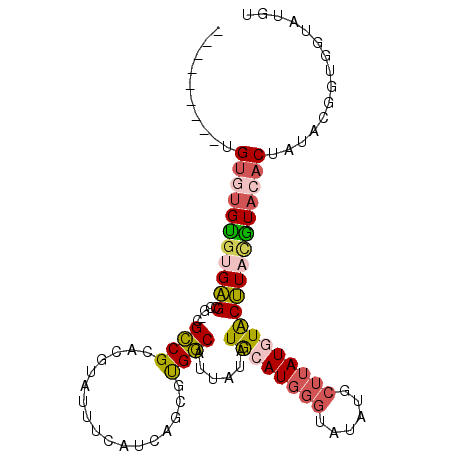
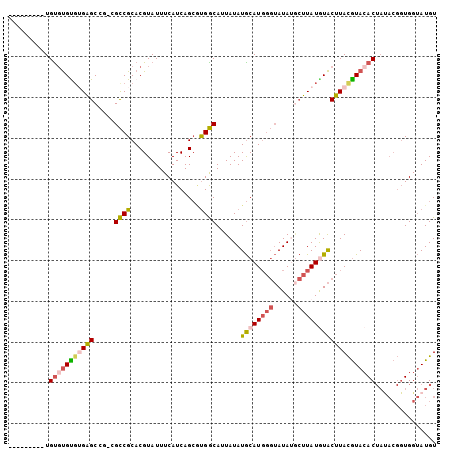
>3L_DroMel_CAF1 10600047 105 - 23771897 ACAUACCACCGUAUAGUGUACGUAAGUAUAUAAGCAUAUACCCAUGCAUAUAAUGCCACGCUGAUGAAAUACGUGCGGCGUCGGCUCACGCAAACACACACACAC .........(((...((((((....))))))..((((......)))).......((((((((((((.....))).)))))).)))..)))............... ( -25.20) >DroSec_CAF1 482437 96 - 1 ACAUACCACCGUAUAGUGUACGUAAGUACAUAAGCAUAUACCCAUGCAUAUAAUGCCACUCUGAUGAAAUACGUGCGGCGACGGCUCUGACACGCA--------- ......(.((((((.((((((....))))))..((((......)))).........................)))))).)...((........)).--------- ( -20.90) >DroSim_CAF1 482497 96 - 1 ACAUACCACCGUAUAGUGUACGUAAGUACAUAAGCAUAUACCCAUGCAUAUAAUGCCACGCUGAUGAAAUACGUGCGGCGACGGCUCACACACACA--------- ..((((....)))).((((((....))))))..((((......)))).......(((.((((((((.....))).)))))..)))...........--------- ( -24.00) >DroEre_CAF1 489739 94 - 1 ACAUACCACCGUAUAGUGUACGUAAGUACAUAAGCGUAUACCCAUGCAUAUAAUGCCAUGCUGAUGAAAUACGUGCGGC---GGCUCUCGCACACA--------C .....((.((((...((((((....))))))..((((((...(((((((........))))..)))..)))))))))).---))............--------. ( -25.10) >DroYak_CAF1 492234 94 - 1 ACAUACCACCGUGUAGUGUACGUAAGUACAUAAGAGUAUACCCAUGCAUAUAAUGCCGUGCUGAUGAAAUACGUGCGGC---GGCUCACACACACA--------C ..........((((.((((((....))))))..(((((((........)))..(((((..(...........)..))))---))))))))).....--------. ( -25.70) >DroAna_CAF1 472698 85 - 1 ACAUCCCA-------GUUUAUAUAUGCAUAUAUUUAUAU----AUGUAUAUAAUGUCAGGCUGAUGAAAUACGUGCGACAGAGGCACACACGCACA--------- .((((((.-------(....((((((((((((....)))----)))))))))....).))..))))......(((((.............))))).--------- ( -20.62) >consensus ACAUACCACCGUAUAGUGUACGUAAGUACAUAAGCAUAUACCCAUGCAUAUAAUGCCACGCUGAUGAAAUACGUGCGGCG_CGGCUCACACACACA_________ ...............((((((....))))))..((((......)))).......(((..(((((((.....))).))))...))).................... (-14.59 = -14.87 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:58 2006