
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,586,481 – 10,586,573 |
| Length | 92 |
| Max. P | 0.790784 |

| Location | 10,586,481 – 10,586,573 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -17.95 |
| Energy contribution | -18.53 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.790784 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10586481 92 + 23771897 AUUUCGCCGCACGAUUGGUAUGUCAUGUCAUGUCAAUCUUU-------CCGGAAUAUUCUCUUCCCGGCG------CCGGGAUUCGUGACCCUUUAGG-------CCAGAGA .((((((((...(((((..(((......)))..)))))...-------..((((.......))))))))(------(((((........)))....))-------)..)))) ( -26.50) >DroSec_CAF1 466034 92 + 1 AUUUCGCCGCACGAUUGGUAUGUCAUGUCAUGUCAAUCUUU-------CCGGAAUAUUCUCUUCCCGGCG------CCGGGAUUCGUGUCCCUUUAGC-------CCAGCGA ...((((.((..(((((..(((......)))..)))))...-------..((.((((.....(((((...------.)))))...)))).))....))-------...)))) ( -27.60) >DroSim_CAF1 468627 92 + 1 AUUUCGCCGCACGAUUGGUAUGUCAUGUCAUGUCAAUCUUU-------CCGGAAUAUUCUCUUCCCGGCG------CCGGGAUUCGUGGCCCUUUAGG-------CCAGCGA ......(((...(((((..(((......)))..)))))...-------.)))..........(((((...------.))))).((((((((.....))-------))).))) ( -31.70) >DroEre_CAF1 475432 96 + 1 AUUUCGCCGCACGAUUGGUAUGUCAUGUCAUGUCAAUCUUU-------CCGGAAUACUCCCAUCCCGGCG--AUUCCCGGGAUUCGUGGUCCUUUAGG-------CCAGAGA ......(((...(((((..(((......)))..)))))...-------.)))....(((..(((((((..--....)))))))...(((((.....))-------)))))). ( -31.10) >DroWil_CAF1 747372 105 + 1 AUUUUGCCGCACGAUUGGCAUGU-----CAUGUCAAUCUUUGUAUGUGCCGCAACCACCCCAUCUGGGCCCCCUUCCCUCCCCUCCUUGCCCUUUACCCUUCUCGGC--AGU ...((((.(((((((((((((..-----.))))))))).......)))).))))....(((....)))..................(((((.............)))--)). ( -24.03) >DroYak_CAF1 477497 92 + 1 AUUUCGCCGCACGAUUGGUAUGUCAUGUCAUGUCAAUCUUU-------CCGGAAUAUUCCCAUCCCGGCG------CCGGGAUUCGUAACCCUUUAGG-------CCAGUGA .....(((....(((((..(((......)))..)))))...-------..((..(((....((((((...------.))))))..)))..))....))-------)...... ( -25.70) >consensus AUUUCGCCGCACGAUUGGUAUGUCAUGUCAUGUCAAUCUUU_______CCGGAAUAUUCCCAUCCCGGCG______CCGGGAUUCGUGGCCCUUUAGG_______CCAGAGA .........(((((((((((((......))))))))))........................((((((........))))))...)))........................ (-17.95 = -18.53 + 0.58)

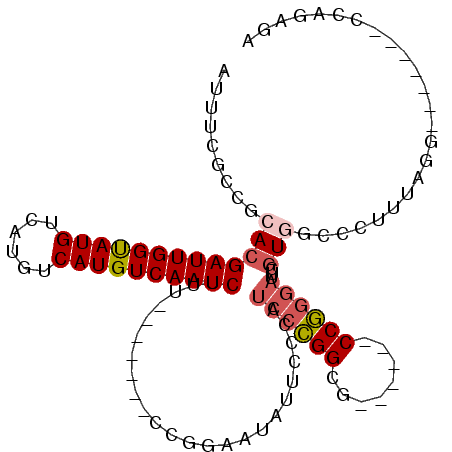
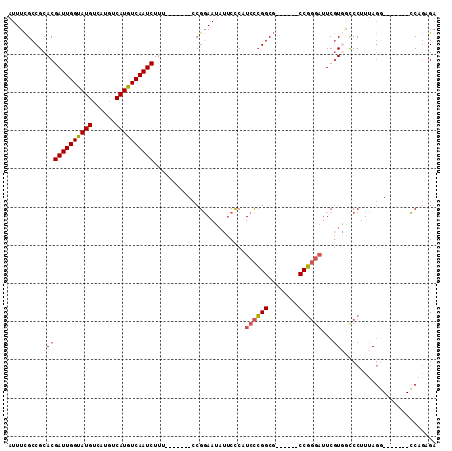
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:54 2006