
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
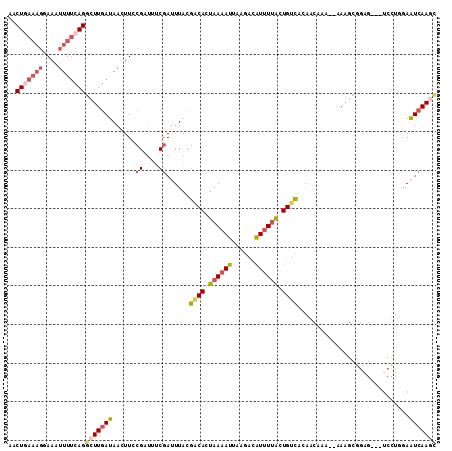
| Location | 10,584,042 – 10,584,148 |
| Length | 106 |
| Max. P | 0.998977 |

| Location | 10,584,042 – 10,584,148 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 69.91 |
| Mean single sequence MFE | -22.53 |
| Consensus MFE | -8.91 |
| Energy contribution | -10.60 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.40 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10584042 106 + 23771897 AGCUUAAAGGAAAUUUUCAGGCUUGAUAACUUACGAUUUCGAUUUACGACACUAAAAUUAAGACGUCUUACUGCCACAGCAAA--AAAGCUGAG---UCCUGGAAUCAACU (((((((((....)))).))))).................(((((..(((........((((....))))......((((...--...)))).)---))...))))).... ( -17.00) >DroSec_CAF1 463506 106 + 1 GACUGAAAGGAAAUUUUCAGGCUUGAUAACUUCCGCUUUCGAUUUACGACACUAAAAUUAAGACAUUUUACUGUCAUAACAUA--AAAGCAGAG---UCUUAGAAUCAAGC ..(((((((....)))))))(((((((.((((..(((((....(((.((((.((((((......)))))).)))).)))....--))))).)))---)......))))))) ( -24.60) >DroSim_CAF1 465875 106 + 1 AACUAAAAGGAAAUUUUCAGGCUUGAUAAUUUCCGAUUUCUAUUUACGACACUAAAAUUAACAUGUUUAACUGUUACAGCAAA--AAAGCGAAG---UCCUGGAAUCAAAC ........(((((((.((......)).)))))))((((((.......(((........(((((........)))))..((...--...))...)---))..)))))).... ( -18.40) >DroEre_CAF1 472933 100 + 1 AGCUG-----AAAUUUGCAGGCUUAAUAACUUUC------GAUUUAUGACAAUAAAAUGAGGGCAUUUUGCUGUCACAACCAAAGAAAGCGGUGAUAUCUGCAAGUCAAGA ..((.-----..(((((((((....((..(((((------......(((((.(((((((....))))))).)))))........)))))..))....)))))))))..)). ( -25.44) >DroYak_CAF1 474965 86 + 1 GACUGAAAGGAAAUUUUCAGGCUUGAUAACUUCCGAUGCCGAUUUACGGCAAUAAAAUUAAGGCAUUUUACUGCCAA-------------------------AAGUCAAGC ..(((((((....)))))))(((((((.........(((((.....)))))..........((((......))))..-------------------------..))))))) ( -27.20) >consensus AACUGAAAGGAAAUUUUCAGGCUUGAUAACUUCCGAUUUCGAUUUACGACACUAAAAUUAAGACAUUUUACUGUCACAACAAA__AAAGCGGAG___UCCUGGAAUCAAGC ..(((((((....)))))))(((((((......((....))......((((.((((((......)))))).)))).............................))))))) ( -8.91 = -10.60 + 1.69)


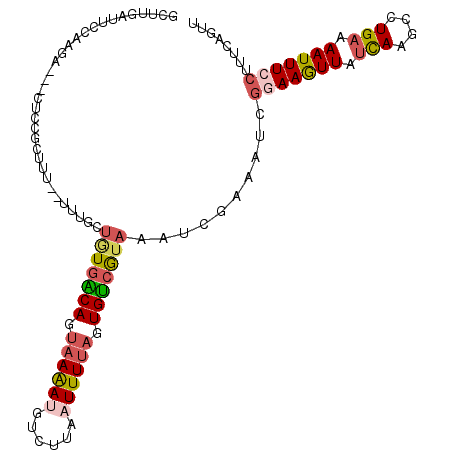
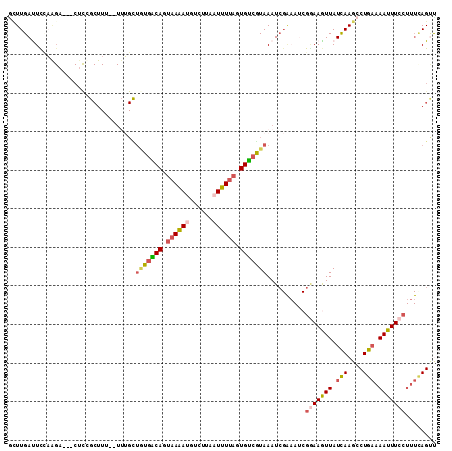
| Location | 10,584,042 – 10,584,148 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 69.91 |
| Mean single sequence MFE | -25.92 |
| Consensus MFE | -12.16 |
| Energy contribution | -13.20 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.47 |
| SVM decision value | 3.31 |
| SVM RNA-class probability | 0.998977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10584042 106 - 23771897 AGUUGAUUCCAGGA---CUCAGCUUU--UUUGCUGUGGCAGUAAGACGUCUUAAUUUUAGUGUCGUAAAUCGAAAUCGUAAGUUAUCAAGCCUGAAAAUUUCCUUUAAGCU ((..((((.((((.---((((((...--...)))).((((.(((((........))))).)))).......((...(....)...)).))))))..))))..))....... ( -18.70) >DroSec_CAF1 463506 106 - 1 GCUUGAUUCUAAGA---CUCUGCUUU--UAUGUUAUGACAGUAAAAUGUCUUAAUUUUAGUGUCGUAAAUCGAAAGCGGAAGUUAUCAAGCCUGAAAAUUUCCUUUCAGUC (((((((.....((---(((((((((--(...((((((((.((((((......)))))).))))))))...)))))))).)))))))))))((((((......)))))).. ( -39.40) >DroSim_CAF1 465875 106 - 1 GUUUGAUUCCAGGA---CUUCGCUUU--UUUGCUGUAACAGUUAAACAUGUUAAUUUUAGUGUCGUAAAUAGAAAUCGGAAAUUAUCAAGCCUGAAAAUUUCCUUUUAGUU ((((((((((..((---(...((...--...))..(((((........)))))........))).......(....))))....)))))))((((((......)))))).. ( -16.40) >DroEre_CAF1 472933 100 - 1 UCUUGACUUGCAGAUAUCACCGCUUUCUUUGGUUGUGACAGCAAAAUGCCCUCAUUUUAUUGUCAUAAAUC------GAAAGUUAUUAAGCCUGCAAAUUU-----CAGCU ..((((.((((((........((((((.....(((((((((.((((((....)))))).)))))))))...------))))))........))))))...)-----))).. ( -25.69) >DroYak_CAF1 474965 86 - 1 GCUUGACUU-------------------------UUGGCAGUAAAAUGCCUUAAUUUUAUUGCCGUAAAUCGGCAUCGGAAGUUAUCAAGCCUGAAAAUUUCCUUUCAGUC ((((((...-------------------------..((((......)))).(((((((..(((((.....)))))...)))))))))))))((((((......)))))).. ( -29.40) >consensus GCUUGAUUCCAAGA___CUCCGCUUU__UUUGCUGUGACAGUAAAAUGUCUUAAUUUUAGUGUCGUAAAUCGAAAUCGGAAGUUAUCAAGCCUGAAAAUUUCCUUUCAGUU .................................(((((((.((((((......)))))).)))))))..........(((((((.(((....))).)))))))........ (-12.16 = -13.20 + 1.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:53 2006