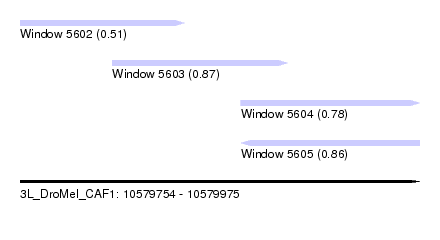
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 10,579,754 – 10,579,975 |
| Length | 221 |
| Max. P | 0.868329 |

| Location | 10,579,754 – 10,579,845 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.36 |
| Mean single sequence MFE | -16.61 |
| Consensus MFE | -11.14 |
| Energy contribution | -11.00 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.67 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10579754 91 + 23771897 AGCUA---CUCUUUCCACAUUCUACUUACCAUUUG------------UGCUCC----G----------CUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGU ((((.---.......((((..............))------------))....----.----------....)))).((....(((..(((((....)))))....))).....)).... ( -15.97) >DroSec_CAF1 459163 90 + 1 AG-UC---CUCUUUCCACAUUCAACUUACCAUUUG------------UGCUCC----G----------CUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGU ..-..---........((((.(((...((...(((------------((((.(----(----------...((((..((.....))..))))....)).))).)))).))..))).)))) ( -14.80) >DroEre_CAF1 468489 101 + 1 AACUC---CGCUUUCCACAUUCAACUUACCAUUUG------------UGCUCC----GCUCCGGUCCGCUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACAUUUUUGCAUGU .....---.((....((((..............))------------))....----))..((((..((........))..))))...(((((....)))))(((((.....)))))... ( -18.24) >DroYak_CAF1 470321 94 + 1 AGCUCCUCCUCUUUCCACAUUCAACUUACCAUUUG------------UGCUCC----G----------CUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGU (((............((((..............))------------))....----(----------(....))..)))........(((((....)))))(((((.....)))))... ( -15.74) >DroAna_CAF1 452390 70 + 1 -------------UCCACUUUCAACUU-CCAUUUG------------------------------------CAGCUCGCUUAUCGUCUGUUGCACACGCAGCUGCAACCUUUUUGCAUGU -------------..............-....(((------------------------------------(((((.((..................))))))))))............. ( -12.27) >DroPer_CAF1 525956 105 + 1 AG-UA---ACAGCUCCACUUUCAACUU-CCAUUUGCAGCUGCCUCUCUGCUUCUCUGG----------CUCCUUUUCGCUUAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGU .(-((---(.(((..............-....((((((((((......((..((..((----------(........)))....))..)).......)))))))))).))).)))).... ( -22.63) >consensus AG_UC___CUCUUUCCACAUUCAACUUACCAUUUG____________UGCUCC____G__________CUCCAGCUCGCUUAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGU ........................................................................................(((((....)))))(((((.....)))))... (-11.14 = -11.00 + -0.14)

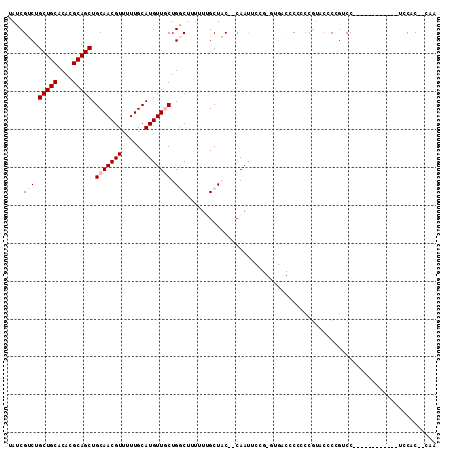
| Location | 10,579,805 – 10,579,902 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10579805 97 + 23771897 UAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUAC--CAAUUCCGGGUGACCCCUCCGUCCACCGUCC------------UCCGC--CAA ........(((((....))))).(((((((......)))))))((((....(((....--)))...(((..(((......)))..)))...------------...))--)). ( -26.20) >DroPse_CAF1 516769 91 + 1 UAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUUCUGACUUUUU-GCUCCAGCAACA---------GCACCCGUACCCGGGCC------------AGCACUCGCA .....((.(((((....)))))(((........(((.((((.(((.......-....))).))))---------)))((((....))))..------------.)))...)). ( -25.50) >DroSec_CAF1 459213 97 + 1 UAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUAC--CAAUUCCGGGUGACCCCUCCGUCCUCCGUCC------------UCCGC--CAA ........(((((....))))).(((((((......)))))))((((....(((....--)))...((((.(((......))).))))...------------...))--)). ( -27.00) >DroYak_CAF1 470375 100 + 1 UAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUU-UUGCUAC--CAAUUCCGUGUGACCCC--------CCUUCCCCCUUUCAUCCACCCCC--CUC ............(((((((((((((((.....))))).)))))((((...-..)))).--.......))))).....--------.......................--... ( -21.80) >DroPer_CAF1 526021 91 + 1 UAUCGGCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUUCUGACUUUUU-GCUCCAGCAACA---------GCACCCGUACCCGGGCC------------UGCACUCGCA .....((.(((((....))))).))........(((.((((.(((.......-....))).))))---------(((((((....))))..------------)))....))) ( -26.50) >consensus UAUCGUCUGCUGCACACGCAGCUGCAACGUUUUUGCAUGUUGCUGGCUUUUUUGCUAC__CAAUUCCG_GUGACCCCCCCGUACCCCGUCC____________UCCAC__CAA ....(((.(((((....))))).(((((((......))))))).))).................................................................. (-12.90 = -13.70 + 0.80)

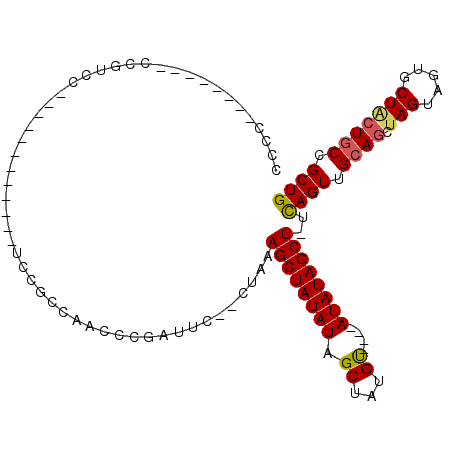
| Location | 10,579,876 – 10,579,975 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 70.89 |
| Mean single sequence MFE | -19.85 |
| Consensus MFE | -14.15 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10579876 99 + 23771897 CCCCUCCGUCCACCGUCC------------UCCGCCAACCCGAUUCCCCUAAAGCUAUAUAGCUAUGUGUAUAUAUAGCU-UCAGUUGCAGCUAGUUGUGCUUGUGCCGCUG ..................------------...((((((..(......((.((((((((((((.....)).)))))))))-).))......)..)))).))........... ( -17.50) >DroSec_CAF1 459284 93 + 1 CCCCUCCGUCCUCCGUCC------------UCCGCCAACCCGAUUC--CUAAAGCUAUAUAGCUAUGU----AUAUAGCU-UCAGUUGGAGCUAGUAGUGCUACUGCCGCUG ..................------------.........((((((.--...((((((((((......)----))))))))-).))))))(((..((((.....)))).))). ( -19.60) >DroSim_CAF1 461648 93 + 1 NNNNNNNNNNNNNNNNNN------------NNNGCCAACCCGAUUC--CUAAAGCUAUAUAGCUAUGU----AUAUAGCU-UCAGUUGCAGCUAGUAGUGCUACUGCCGCUG ..................------------................--...((((((((((......)----))))))))-)((((.((((.(((.....))))))).)))) ( -21.00) >DroYak_CAF1 470445 98 + 1 CCCC--------CCUUCCCCCUUUCAUCCACCCCCCUCCCCAAUUC--CCAAAGCUAUAUAGCUAUGC----AUAUAGCUUCUAGUUGCAGCUAGUAGUGCUGCUGCCGCUG ....--------..................................--...(((((((((.((...))----))))))))).((((.(((((.((.....))))))).)))) ( -21.30) >consensus CCCC________CCGUCC____________UCCGCCAACCCGAUUC__CUAAAGCUAUAUAGCUAUGU____AUAUAGCU_UCAGUUGCAGCUAGUAGUGCUACUGCCGCUG ....................................................((((((((.((...))....))))))))..((((.((((.(((.....))))))).)))) (-14.15 = -14.40 + 0.25)

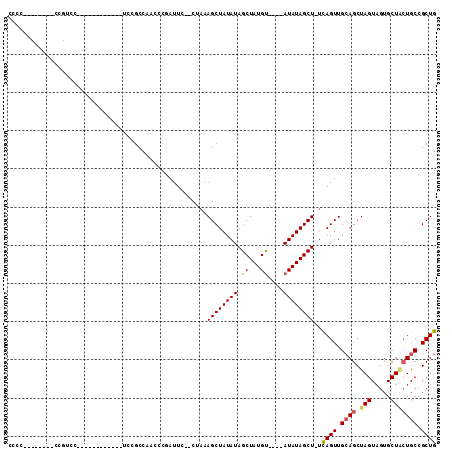
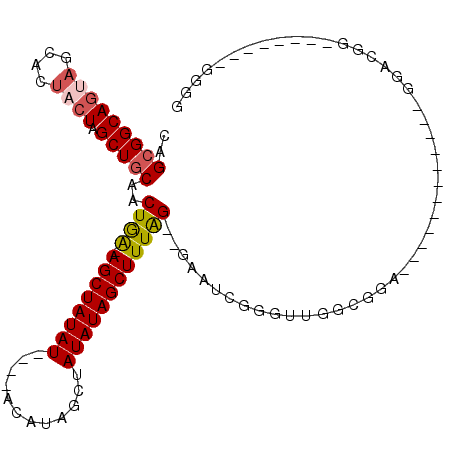
| Location | 10,579,876 – 10,579,975 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 70.89 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -15.31 |
| Energy contribution | -16.00 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 10579876 99 - 23771897 CAGCGGCACAAGCACAACUAGCUGCAACUGA-AGCUAUAUAUACACAUAGCUAUAUAGCUUUAGGGGAAUCGGGUUGGCGGA------------GGACGGUGGACGGAGGGG ((.((.(....((.(((((.....(..((((-(((((((((..........)))))))))))))..).....)))))))...------------.).)).)).......... ( -29.90) >DroSec_CAF1 459284 93 - 1 CAGCGGCAGUAGCACUACUAGCUCCAACUGA-AGCUAUAU----ACAUAGCUAUAUAGCUUUAG--GAAUCGGGUUGGCGGA------------GGACGGAGGACGGAGGGG ..((.......)).(..(((((((...((((-((((((((----(......)))))))))))))--.....)))))))..).------------...((.....))...... ( -27.20) >DroSim_CAF1 461648 93 - 1 CAGCGGCAGUAGCACUACUAGCUGCAACUGA-AGCUAUAU----ACAUAGCUAUAUAGCUUUAG--GAAUCGGGUUGGCNNN------------NNNNNNNNNNNNNNNNNN ..(((((((((....)))).)))))..((((-((((((((----(......)))))))))))))--................------------.................. ( -27.30) >DroYak_CAF1 470445 98 - 1 CAGCGGCAGCAGCACUACUAGCUGCAACUAGAAGCUAUAU----GCAUAGCUAUAUAGCUUUGG--GAAUUGGGGAGGGGGGUGGAUGAAAGGGGGAAGG--------GGGG ..((....))..(((..((..((.(((((.((((((((((----((...)).)))))))))).)--)..))).))..))..)))................--------.... ( -25.50) >consensus CAGCGGCAGUAGCACUACUAGCUGCAACUGA_AGCUAUAU____ACAUAGCUAUAUAGCUUUAG__GAAUCGGGUUGGCGGA____________GGACGG________GGGG ..(((((((((....)))).)))))..((((.((((((((............))))))))))))................................................ (-15.31 = -16.00 + 0.69)

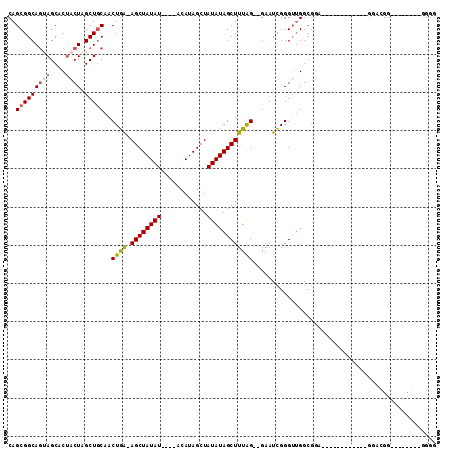
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:01:49 2006